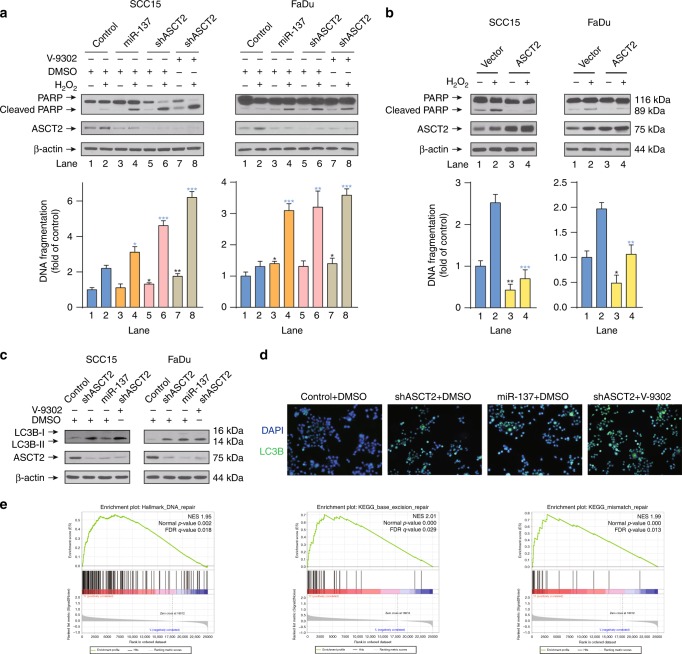
Fig. 3.
ASCT2 inhibits apoptosis and autophagy in HNSCC. a SCC15 and FaDu cells infected with a lentiviral vector expressing ASCT2 shRNA or a control vector were treated with either DMSO or V-9302 (25 µM, 48 h), followed by western blot detection of PARP cleavage and a quantitative apoptosis ELISA. b SCC15 and FaDu cells transfected with an ASCT2 construct or control vector and treated with or without 1 mM H2O2 for 24 h were lysed for the detection of PARP cleavage by western blotting. The results were also verified by a quantitative apoptosis ELISA. c, d SCC15 and FaDu cells transfected with ASCT2 shRNA or a control vector were exposed to DMSO or V-9302 (25 µM, 48 h). The expression of LC3B-II was evaluated by c western blotting, and LC3B-positive cells were detected by d immunofluorescence (images of SCC15 cells are shown). e Patients in the TCGA HNSCC project were divided into two groups (top 25% with high ASCT2 expression and bottom 25% with low ASCT2 expression). GSEA results show that ‘HALLMARK_DNA_REPAIR’, ‘KEGG_BASE_EXCISION_REPAIR’ and ‘KEGG_MISMATCH_REPAIR’ were differentially enriched in the high ASCT2 expression HNSCC patients. ‘*’, ‘**’, ‘***’ and ‘****’ represent ‘p < 0.05′, ‘p < 0.01′, ‘p < 0.001′ and ‘p < 0.0001′, respectively (‘*’ was coloured black with reference to the line 1, ‘*’ was coloured blue with reference to the line 2)