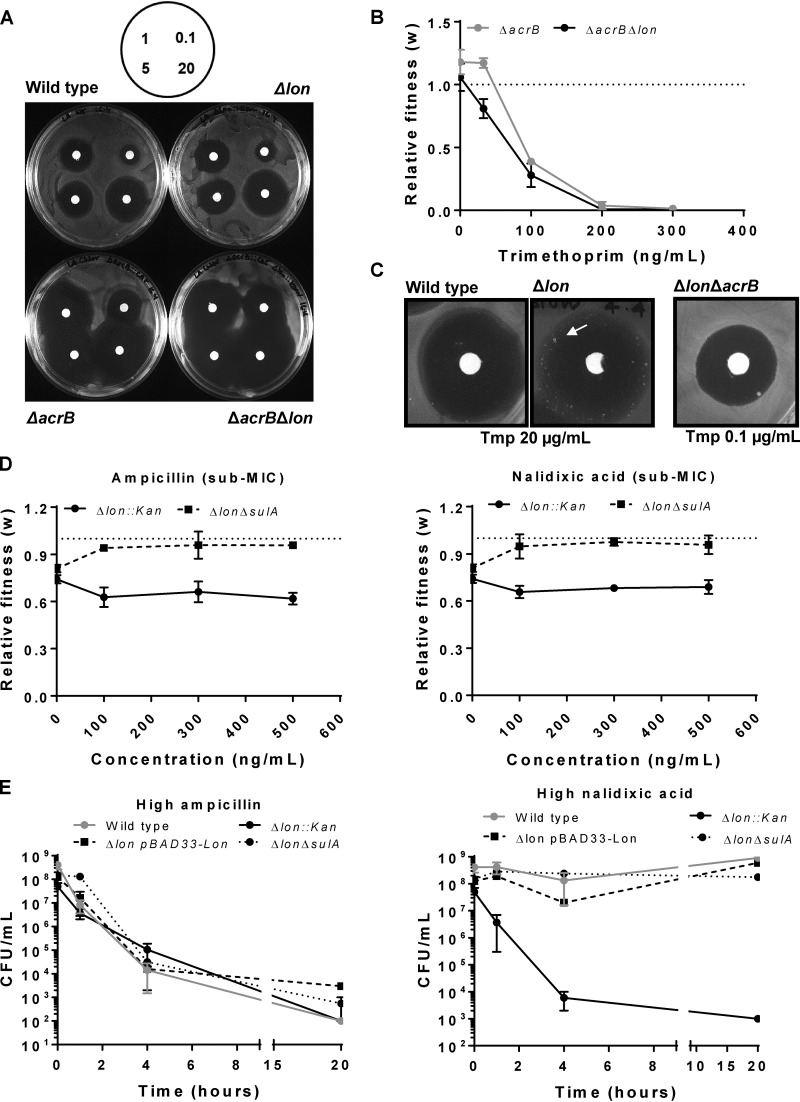
FIG 3.
Trimethoprim tolerance of Lon-deficient E. coli at sub-MICs is dependent of AcrAB-mediated drug efflux. (A) Trimethoprim resistance of indicated genotypes tested by a disc diffusion assay. Concentrations of trimethoprim used in the antibiotic discs are indicated in micrograms per milliliter in the schematic. Representative data from experiments performed twice are shown. (B) Relative fitness (w) of E. coli ΔacrB strain or E. coli ΔacrB Δlon strain compared to wild-type E. coli calculated as a function of trimethoprim concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (C) TDtest showing trimethoprim-tolerant bacteria (arrow) in E. coli Δlon but not in wild-type or ΔacrB Δlon strains. The concentration of trimethoprim used for E. coli ΔacrB Δlon was lower than wild type and Δlon to obtain comparable ZOC diameters. Representative data from experiments performed at least thrice are shown. (D) Relative fitness (w) of E. coli Δlon or E. coli ΔsulA Δlon strains compared to wild-type E. coli calculated as a function of ampicillin or nalidixic acid concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (E) Survival of wild type and indicated mutants of E. coli at a high concentration of ampicillin (20 μg/ml) or nalidixic acid (50 μg/ml). Means ± SEM of results from 3 independent experiments are plotted.