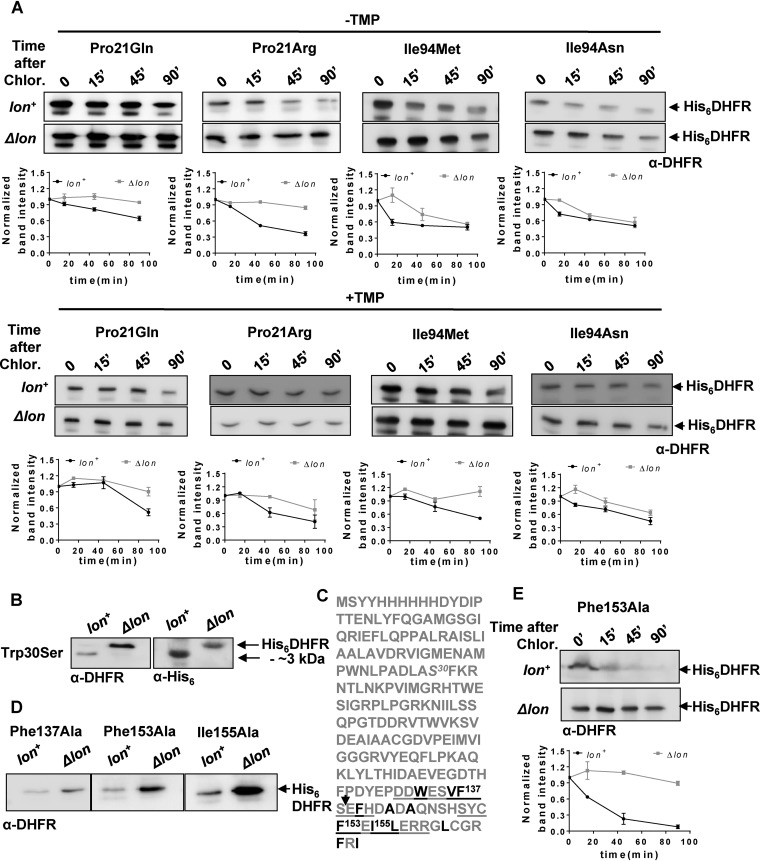
FIG 6.
In vivo proteolytic stabilities explain disparities between trimethoprim-resistant phenotypes of DHFR mutants in lon+ and Δlon backgrounds. (A) In vivo stabilities of plasmid-encoded mutants of DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed using a chloramphenicol chase shutoff assay. After inhibition of protein synthesis by addition of chloramphenicol, levels of DHFR were assessed using immunoblotting with DHFR-specific polyclonal antibody (α-DHFR) at the indicated times in the presence or absence of trimethoprim in the growth medium. Representative immunoblot data from experiments performed at least thrice are shown. Below each blot, quantitation of band intensities normalized to intensity at 0 min is plotted as means ± SD of results from 3 independent experiments. (B) Clipping of Trp30Ser DHFR in lon+ E. coli. DHFR expression in lysates of lon+ or Δlon E. coli harboring plasmid coding for His6-DHFR Trp30Ser was analyzed by immunblotting. The clipped fragment of Trp30Ser DHFR was roughly 3 kDa smaller than full-length Trp30Ser DHFR and showed immunoreactivity with anti-DHFR polyclonal and anti-His monoclonal antibodies, indicating clipping at the C terminus of the protein. Due to the relatively lower level of immunoreactivity with anti-His antibody, lysates in which DHFR was overproduced by induction with IPTG were used for immunoblots performed with this antibody. (C) Sequence of plasmid-encoded Trp30Ser DHFR. The Trp30Ser mutation is indicated in italics. The 2 buried β-sheets at the C terminus of DHFR are underlined, and hydrophobic residues are shown in black. Phe137, Ph153, and Ile155, all of which are important for folding of DHFR, are indicated. The expected proteolytic site that would result in an approximately 3-kDa-smaller fragment is indicated by an arrow. (D) Levels of plasmid-encoded Phe137Ala, Phe153Ala, and Ile155Ala mutant DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed by immunoblotting using DHFR polyclonal antibody (α-DHFR). Representative data from 3 independent experiments are shown. (E) In vivo stabilities of plasmid-encoded Phe153Ala DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed using a chloramphenicol chase assay. After inhibition of protein synthesis by addition of chloramphenicol, levels of DHFR were assessed using immunoblotting with DHFR-specific polyclonal antibody (α-DHFR) at the indicated times. Representative data from experiments performed thrice are shown.