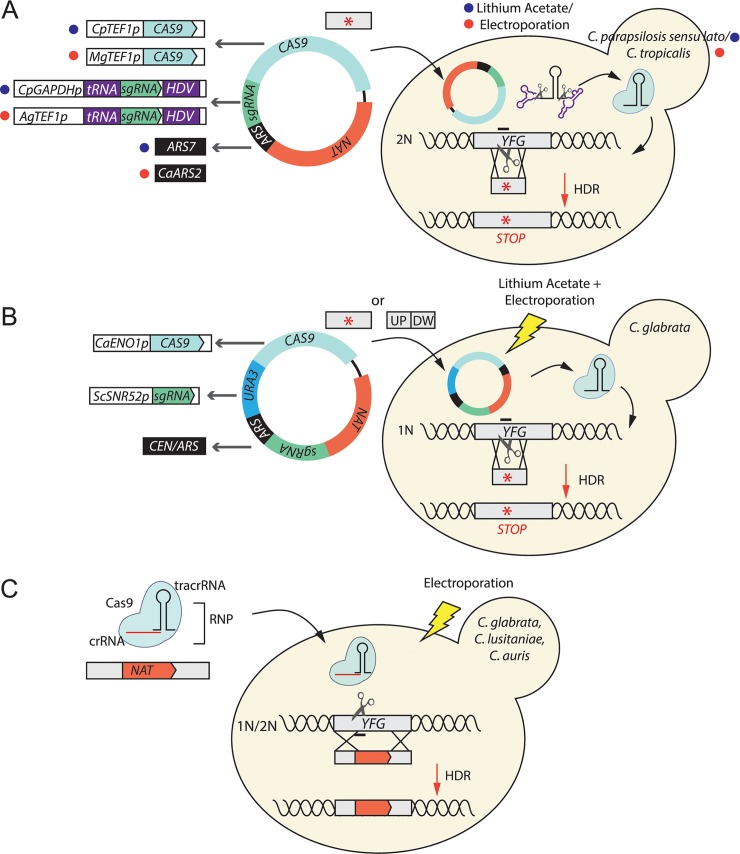
Fig 2. CRISPR–Cas9 methods in non-albicans Candida species.
The vignettes illustrate representative methods for implementation of CRISPR–Cas9 in non-albicans species, including C. parapsilosis sensu lato, C. tropicalis, C. glabrata, C. lusitaniae, and C. auris. The ploidy of the species is indicated next to the DNA helices. (A) Two different selectable episomal plasmids allow the scarless introduction of premature stop codons by HDR in C. parapsilosis sensu lato and C. tropicalis. The relevant species-specific promoters and ARSs are depicted on the left side (blue dot: C. parapsilosis sensu lato; red dot: C. tropicalis). The sgRNA is expressed from an RNA pol II promoter, and the release of the mature molecule is mediated by the cleavage of the upstream tRNA molecule and the downstream HDV ribozyme. The plasmids are easily lost in the absence of selection [29]. (B) A similar approach can be used for gene editing and deletion in C. glabrata. In this case, the sgRNA is expressed from the Saccharomyces cerevisiae RNA pol III SNR52 promoter [30]. (C) The use of CRISPR RNPs, in which the crRNA and tracrRNA are assembled in vitro with Cas9 and then electroporated into the cells, results in the integration of a NAT cassette into the targeted gene in C. glabrata, C. lusitaniae, and C. auris [25]. Although this method does not require species-specific adaptation of the regulatory elements necessary for the expression of the CRISPR elements, marker recycling is not possible. Ag, Ashbya gossypii; ARS, autonomously replicating sequence; Ca, Candida albicans; Cp, C. parapsilosis; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR-RNA; DW, downstream; HDR, homology-directed repair; HDV, human hepatitis delta virus; Mg, Meyerozyma guilliermondii; RNP, RNA–Cas9 protein complex; Sc, Saccharomyces cerevisiae; sgRNA, single-guide RNA; tracrRNA, trans-activating RNA; UP, upstream; YFG, your favorite gene.