Fig 5. Ngfra signaling regulates NSC plasticity independent of Il4ra signaling.
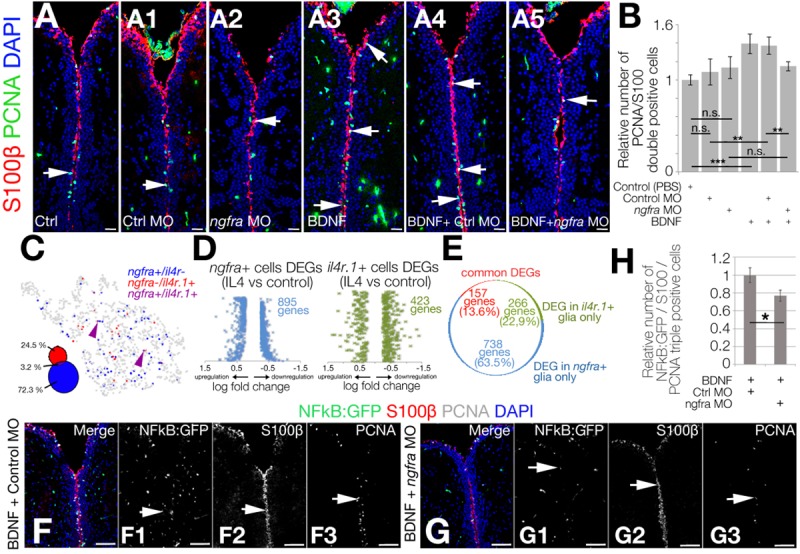
(A–A5) IHC for S100β and PCNA in control (A), control morpholino-injected (A1), ngfra morpholino-injected (A2), BDNF-injected (A3), BDNF + control morpholino-injected (A4), and BDNF + ngfra morpholino-injected (A5) brains. (B) Quantification for the relative number of proliferating glial cells. (C) Co-representation of ngfra and ilr4 expressions on glial tSNE plot. (D) DEG plots for ngfra-positive and il4r-positive neural stem cells after IL4 treatment. (E) Pie chart distribution of unique DEGs. Note that the overlapping genes constitute only 13.6% of all DEGs. (F, G) IHC for S100β, NFkB-driven GFP, and PCNA in BDNF + control morpholino-injected (F–F3) and BDNF + ngfra morpholino-injected brains (G–G3).(H) Quantification graph for relative numbers of proliferating stem cells with active NFkB signaling. Scale bars equal 100 μM. Data are represented as mean ± SEM. See also S10 Fig. See S5 Data, S6 Data, and S7 Data for supporting information. BDNF, brain-derived neurotrophic factor; DEG, differentially expressed gene; GFP, green fluorescent protein; IHC, immunohistochemistry; IL4, interleukin-4; NFkB, nuclear factor 'kappa-light-chain-enhancer' of activated B-cells; Ngfr, nerve growth factor receptor; NSC, neural stem cell; PCNA, proliferating nuclear cell antigen; S100β, S100 calcium-binding protein B; tSNE, t-Distributed Stochastic Neighbor Embedding.
