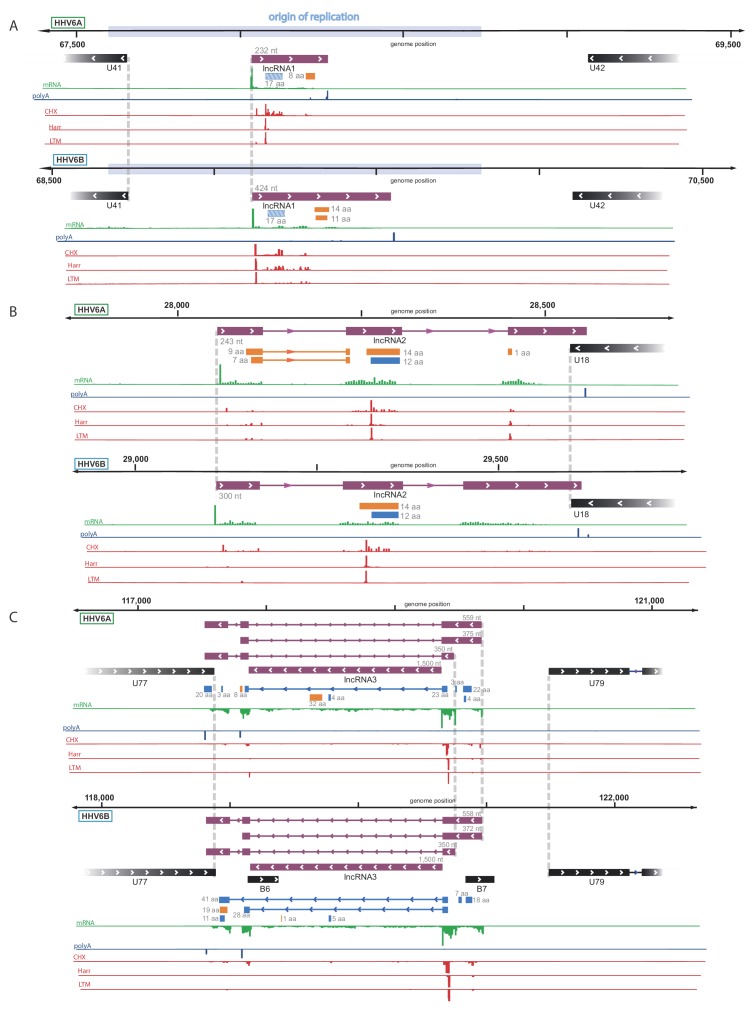
Figure 4. Identification of three highly abundant and conserved viral long non-coding RNAs (lncRNAs).
Viral transcripts that appear to be lncRNAs are shown as purple rectangles. Reads from RNA-seq are presented in green and reads containing polyA are presented in blue. The ribosome profiling (CHX), Harringtonine (Harr) and lactimidomycin (LTM) profiles are presented in red. (A) A transcript initiating within the origin of replication. One putative ORF not detected by our predictions (see Figure 6) is shown as a striped blue rectangle. (B) A spliced transcript initiating between U17 and U18. (C) Three possible isoforms of a spliced transcript with alternative splicing, initiation and termination, as well as a putative stable intron.
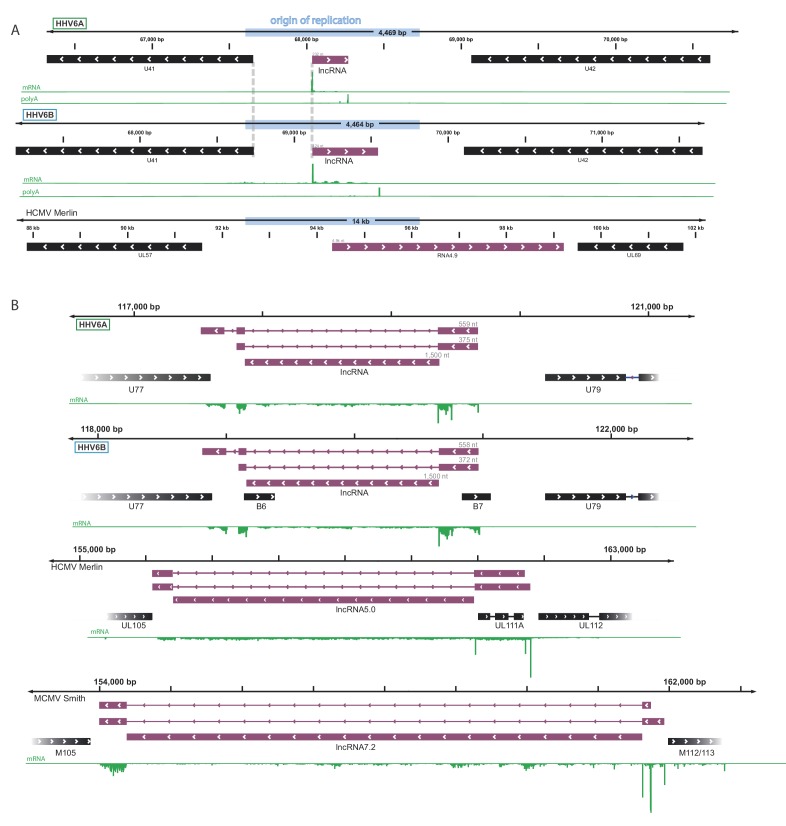
Figure 4—figure supplement 1. Conservation by synteny of newly discovered HHV-6 lncRNAs.
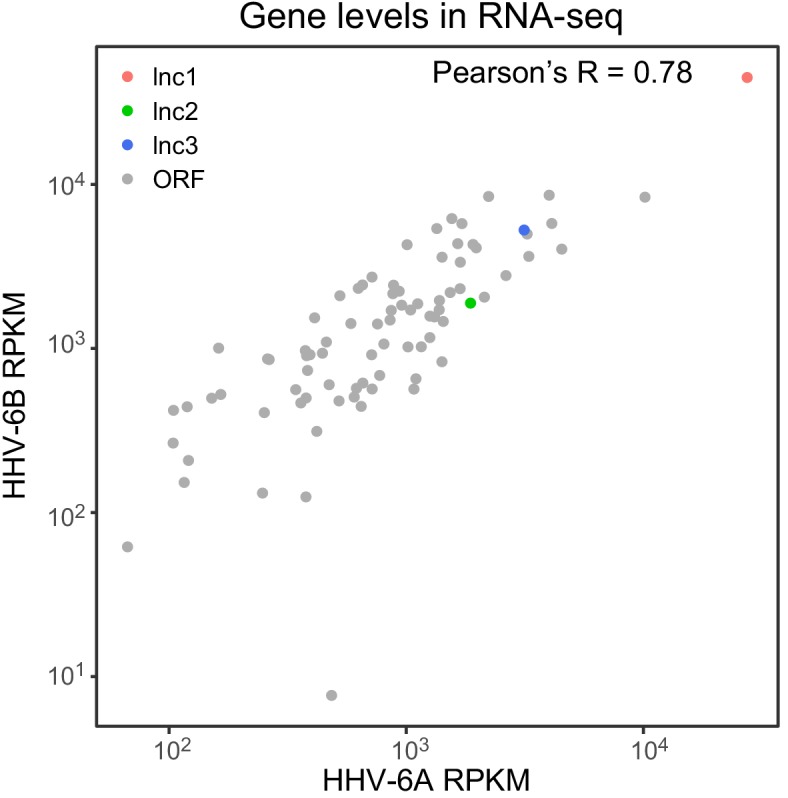
Figure 4—figure supplement 2. RNA abundance of canonical ORFs and viral lncRNAs is conserved between HHV-6A and HHV-6B.