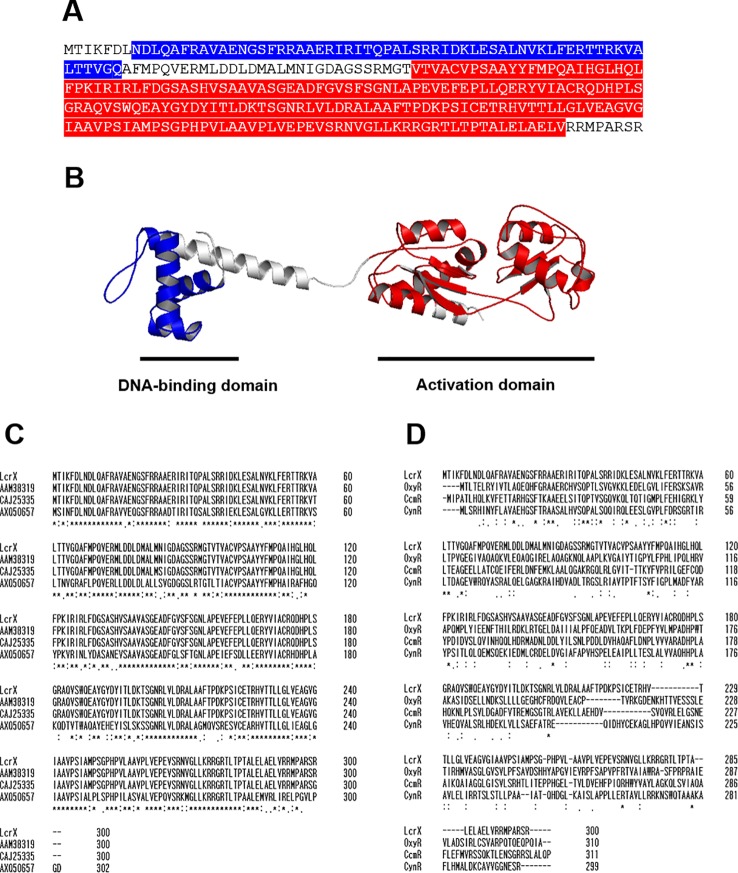
Figure 1.
Predicted three-dimensional structure and sequence alignment of LcrX. (A) Deduced amino acid sequence of LcrX where blue and red boxes indicate DNA-binding domain and LysR activation domain, respectively. (B) Predicted 3D structure obtained using the protein modeling server I-TASSER and PyMOL program. (C) Comparison of amino acid sequences of LcrX with its homologs in Xanthomonas spp. and Stenotrophomonas spp. using the ClustalOmega program. CAJ25335 from X. campestris pv. vesicatoria str. 85-10, AAM38319 from X. citri pv. citri str. 306, AXQ50657 from Stenotrophomonas rhizophila str. GA1 (D) Comparison of amino acid sequences of LcrX with LysR type transcriptional regulators whose functions were previously characterized. OxyR from Pseudomonas aeruginosa str. PAO1, CcmR from Synechococcus sp. str. PCC 7002, and CynR from Escherichia coli str. K-12. ‘*’, ‘.’, and ‘:’ indicate most conserved residues, semi-conserved sequence, respectively.