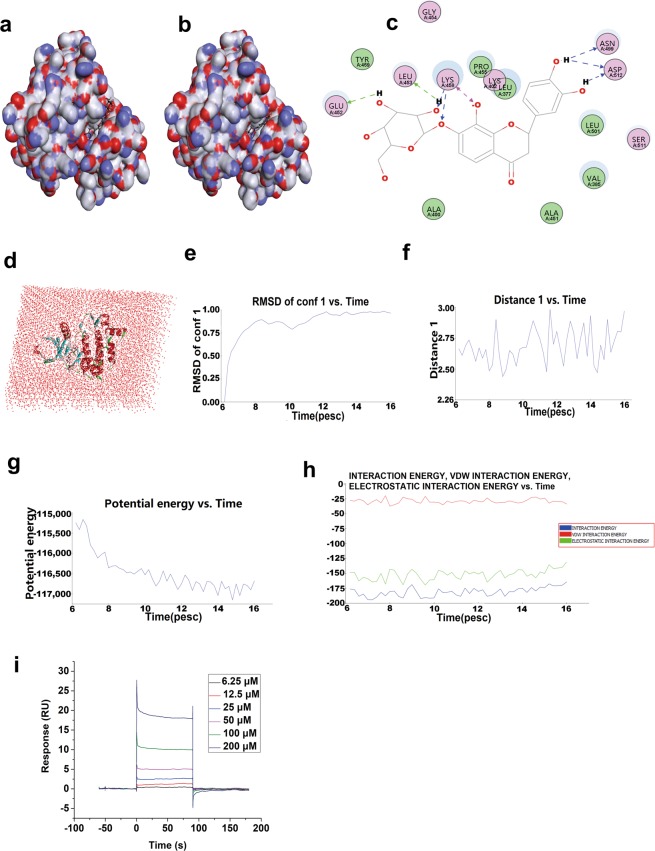
Figure 4.
Protein-ligand interactions, molecular dynamics and binding affinity analysis of Syk and FM. (a) Interaction models of Syk and FM in the optimal docking pose. The -CDOCKER_INTERACTION_ENERGY score was −75.1069 kcal/mol. (b) Interaction models of Syk and ligand LASW836 in the optimal docking pose. The -CDOCKER_INTERACTION_ENERGY score was −57.4404 kcal/mol. (c) Detailed interaction modes of Syk and FM in the optimal docking pose. (d) Model of the Syk-FM complex in solvent. (e) Drug positional RMSD. (f) Distance between O of FM and HN in the amino residue of Lys458 in Syk. (g) Potential energy of the amino residue group between Syk and FM. (h) Interaction energy of the amino residue group between Syk and FM evaluated using molecular dynamics. (i) Real-time binding affinity measurements of FM using Biacore T200. Representative sensorgrams obtained from injection of different concentrations of FM (6.25, 12.5, 25, 50, 100, and 200 μM; curves from bottom to top) over the immobilized Syk surface on the CM5 chip. Note: FM is displayed in the stick representation while residues of Syk are presented as balls. Water is depicted in pink.