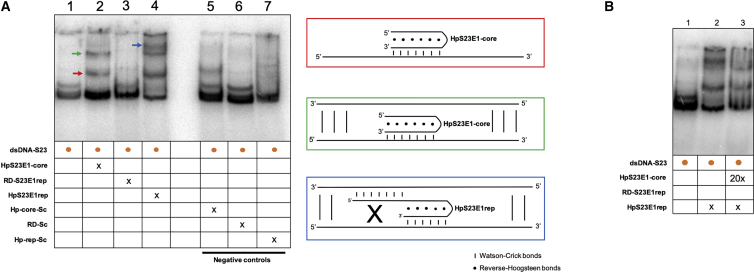
Figure 5.
Binding of the Repair-PPRH to Its Target Sequence
Gel-shift assays using a 160-bp 32P-radiolabeled dsDNA probe (dsDNA-S23) containing the mutation present in the S23 mutant and its flanking regions. The unlabeled oligodeoxynucleotides present in each binding reaction are indicated. (A) Lane 1, dsDNA-S23 probe alone; lane 2, dsDNA-S23 plus HpS23E1-core (100 nM); lane 3, dsDNA-S23 plus RD-S23E1rep (100 nM); lane 4, dsDNA-S23 plus HpS23E1rep repair-PPRH (100 nM); lane 5, dsDNA-S23 plus Hp-core-Sc (100 nM); lane 6, dsDNA-S23 plus RD-Sc (100 nM); lane 7, dsDNA-S23 plus Hp-rep-Sc (100 nM). Color arrows indicated the different molecular species that are generated upon incubation with the different oligodeoxynucleotides. The color of the arrows matches those of the panels corresponding to the proposed structures shown on the right panel. (B) Competition assay. Lane 1, dsDNA-S23 probe alone; lane 2, dsDNA-S23 probe plus HpS23E1rep repair-PPRH (100 nM); lane 3, dsDNA-S23 probe plus HpS23E1rep (100 nM) competed with HpS23E1-core (2 μM). 20x indicates that the oligonucleotide was added in a concentration 20 times greater than x.