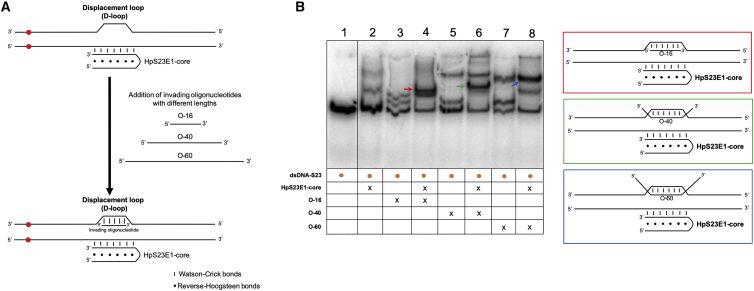
Figure 6.
Strand Displacement upon Binding of the Repair-PPRH to the Target Sequence
(A) Schematic representation of the strategy used to study the formation of a displacement-loop (D-loop) structure. The hairpin core (HpS23E1-core) of the repair-PPRH was incubated with the dsDNA-S23 probe, and then different oligonucleotides varying in length and complementary to the displaced strand were added to the binding reaction. (B) Gel-shift assays using a 160-bp 32P-radiolabeled dsDNA probe (dsDNA-S23) containing the mutation in S23 cells and its flanking regions. In all cases, an amount of 60 ng of each one of the unlabeled oligodeoxynucleotides was added to the binding reaction. Lane 1, dsDNA-S23 probe alone; lane 2, dsDNA-S23 plus HpS23E1-core; lane 3, dsDNA-S23 plus O-16; lane 4, dsDNA-S23 plus HpS23E1-core plus O-16; lane 5, dsDNA-S23 plus O-40; lane 6, dsDNA-S23 plus HpS23E1-core plus O-40; lane 7, dsDNA-S23 plus O-60; lane 8, dsDNA-S23 plus HpS23E1-core plus O-60. Color arrows indicate the different molecular species that are generated upon incubation with the different oligodeoxynucleotides. The colors of the arrows in the gel image match those of the panels corresponding to the proposed structures shown on the right of the figure.