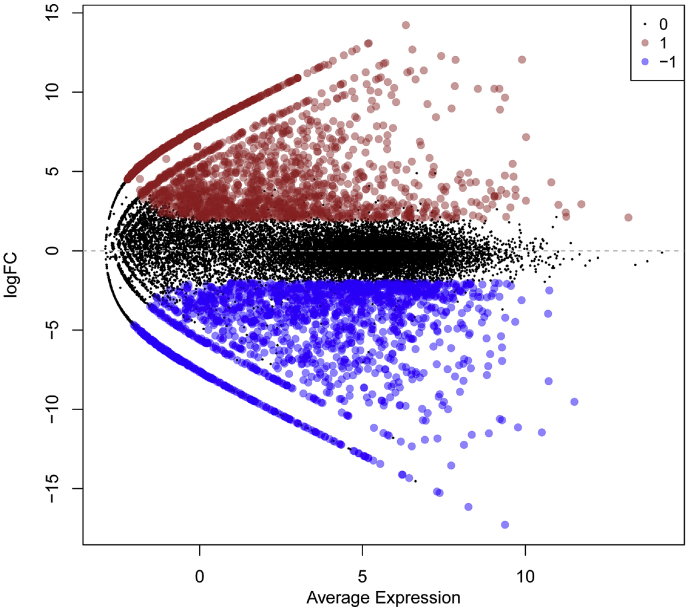
Fig. 2.
The significantly dysregulated lncRNAs in breast cancer cells when compared to normal breast cells (p < 0.05 and fold change ≥ 2.0). Distribution of significantly differentially expressed non-protein coding transcripts (lincRNAs, antisense RNAs, miRNAs, snoRNAs, snRNAs and miscRNAs) obtained from SRA dataset, identified using edgeR (in colour) and annotated according to hg19 fasta file. Mean centered genes were displayed in black colour.