Figure 1.
Knockdown Activity and Cytotoxic Potential in HeLa Cells of 16-nt Gapmers Targeted to HIF1A
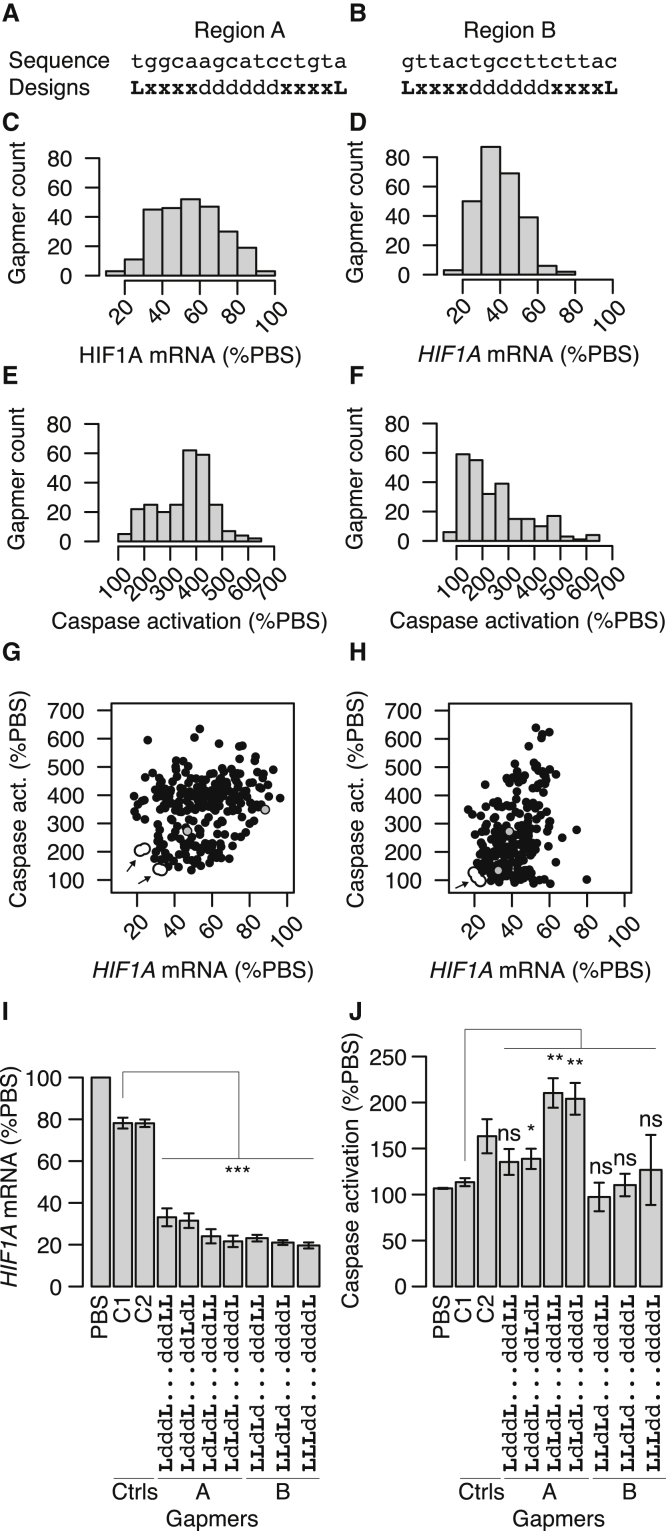
(A and B) Base sequence and LNA/DNA designs for 256 16-nt-long gapmers targeted to region A (A) and region B (B). L, LNA; d, DNA; x, LNA or DNA. (C and D) Histogram of distribution of knockdown activities for the 256 gapmers targeted to region A (C) and region B (D), quantified as HIF1A mRNA levels relative to PBS control after incubation with 5 μM. (E and F) Histogram of distribution of cytotoxic potentials for the 256 gapmers targeted to region A (E) and region B (F), measured by activation of caspase relative to PBS control after transfection of 100 nM. (G and H) Scatterplot of knockdown activity and cytotoxic potential in region A (G) and region B (H). Gray circles indicate the standard 3-10-3 and 4-8-4 designs. Arrows pointing to white circles indicate the top three or four most active and well-tolerated gapmer hits. (I and J) Barplots of knockdown activities of gapmer hits from each region (I), and similarly barplots of cytotoxic potentials (J). Error bars indicate 1 SD. C1 and C2 are negative control gapmers. The variable flank region designs of selected gapmers is shown, with the DNA-only gap region abbreviated to “….” Significance evaluated by Welch t test relative to C1. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.