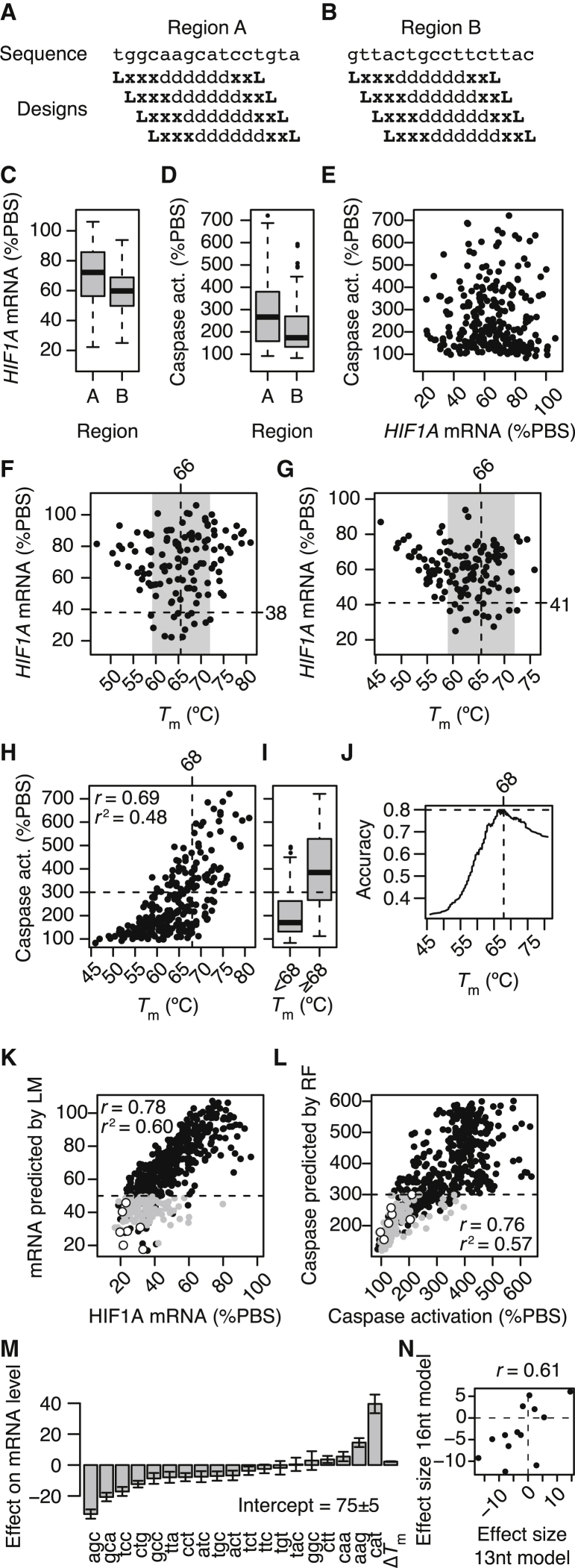
Figure 4.
Evaluation of Tiled 13-nt Gapmers and Prediction of 16-nt Gapmer Properties
(A and B) Base sequence and LNA/DNA designs for 4 × 32 = 128 gapmers, 13 nt in length, tiled within region A (A) and region B (B). L, LNA; d, DNA; x, LNA or DNA. (C) Histograms of distributions of knockdown activities for the 2× 128 13-nt gapmers targeted to region A and B, respectively. Quantified as HIF1A mRNA levels relative to PBS control after incubation with 5 μM. (D) Histograms of distributions of cytotoxic potentials for the 256 gapmers targeted to region A and B, respectively. Quantified by activation of caspase relative to PBS control after transfection of 100 nM. (E) Scatterplot of knockdown activity and cytotoxic potential in both regions A and B. (F and G) Knockdown activity in HeLa cells as a function of Tm for the 128 13-nt gapmers targeting region A (F) and the 128 13-nt gapmers targeting region B (G). Gray area indicates Tm range for the top 10% most active gapmers. Vertical dashed line indicates midpoint of this Tm range, and horizontal dashed line indicates cutoff for the top 10% most active gapmers. (H) Caspase activation in HeLa cells as a function of Tm for all 256 13-nt gapmers targeting region A and B. r indicates Pearson’s correlation, r2 indicate coefficient of determination. Horizontal dashed line at 300% indicates suggested cutoff. Vertical dashed line indicates Tm cutoff achieving best classification accuracy. (I) Boxplot showing distributions of caspase activation levels for gapmers divided into a low/mid-Tm group and a high-Tm group based on the optimal Tm cutoff at 68°C. (J) Classification accuracy of gapmers divided into low and high cytotoxic potential (below or above 300%) when varying the Tm threshold. (K) Scatterplot of measured versus predicted mRNA levels for 16-nt gapmers. Predictions made by robust linear model based on 3-mer motifs and ΔTm trained from 13-nt gapmers. (L) Scatterplot of measured versus predicted caspase activation levels for 16-nt gapmers. White dots indicate the most active and well-tolerated gapmers identified, and gray dots indicate 16-nt gapmers predicted to be both active and reasonably well-tolerated. Predictions made by random forest model based on dinucleotide motifs and ΔTm trained from 13-nt gapmers. (M) Fitted effects on mRNA levels of 3-mer motifs in the gap regions, as well as ΔTm for the 13-nt gapmer activity model, the Tm distance from the region-specific optimal Tm, as determined by iterated re-weighted least-squares fitting of a robust linear model. Error bars indicate 1 SD. (N) Correlation between effect sizes identified from fitting the 13-nt gapmers with those from the fitting of 16-nt gapmers.