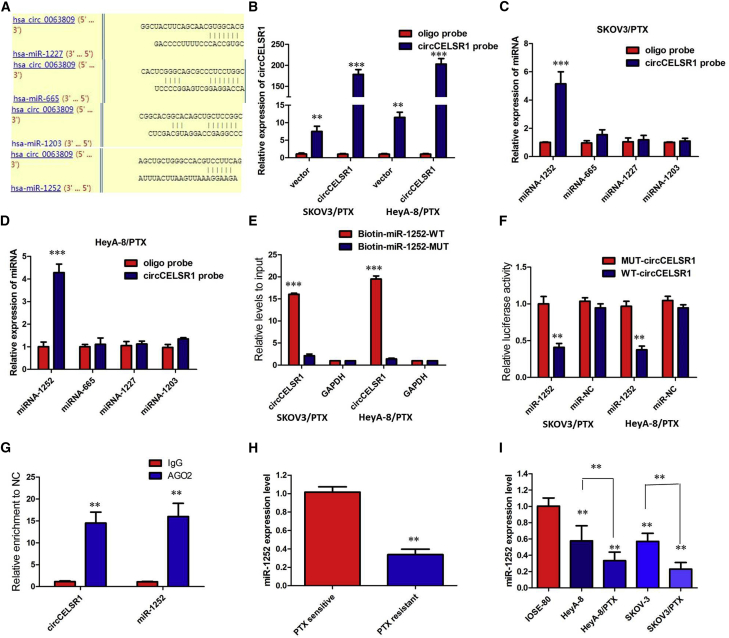
Figure 4.
circCELSR1 Functioned as a Molecular Sponge of miR-1252 in Ovarian Cancer Cells
(A) StarBase v2.0 results showing the sequence of circCELSR1 with highly conserved putative four miRNAs (miRNA-665, miRNA-1227, miRNA-1252, and miRNA-1203) binding sites. (B) Quantitative real-time RT-PCR analysis of circCELSR1 expression in lysates of SKOV3/PTX and HeyA-8/PTX cells with circCELSR1 overexpression following a biotinylated circMAT2B pull-down assay. (C) Expression of four candidate miRNAs was quantified by quantitative real-time PCR after the biotinylated circCELSR1 pull-down assay in SKOV3/PTX cells. miR-1252 was pulled down by circCELSR1 in SKOV3/PTX cells. (D) Expression of four candidate miRNAs was quantified by quantitative real-time PCR after the biotinylated circCELSR1 pull-down assay in HeyA-8/PTX cells. miR-1252 was pulled down by circCELSR1 in HeyA-8/PTX cells. (E) Biotinylated wild-type/mutant miR-1252 was transfected into SKOV3/PTX and HeyA-8/PTX cells with circCELSR1 overexpression. After streptavidin capture, circCELSR1 expression was detected by quantitative real-time PCR. (F) Luciferase activity in SKOV3/PTX and HeyA-8/PTX cells co-transfected with luciferase reporters containing circCELSR1 sequences with wild-type or mutated miR-1252 binding sites and mimics of miR-1252 or controls. (G) circCELSR1 and miR-1252 simultaneously existed in the production precipitated by anti-AGO2. (H) Relative expression of miR-1252 in PTX-resistant ovarian cancer and PTX-sensitive ovarian cancer. (I) Relative expression of miR-1252 in a panel of ovarian cancer cell lines. All tests were at least performed three times. Data are expressed as mean ± SD. **p < 0.01, ***p < 0.001.