Abstract
Background: Radiation resistance poses a major clinical challenge in treatment of nasopharyngeal carcinoma (NPC). Studies have shown that the abnormal expression of microRNAs (miRNAs) is associated with radiosensitivity, however, the mechanisms have not been fully elucidated. The aim of this study, therefore, was to investigate whether ectopic expression of miR-124 is correlated with radiosensitivity in NPC. Methods: In this study, the expression level of miR-124 was evaluated in NPC cell lines and patient specimens using quantitative reverse transcription-PCR (Real-time qPCR). Cell radiosensitivity was determined by colony formation assay. Target prediction algorithms and luciferase assay were used to confirm the target of miR-124. Tumor xenograft model was performed to understand the functions of miR-124 in vivo. Results: We found that miR-124 was down-regulated in both NPC specimens and NPC cell lines. Ectopic expression of miR-124 increased radiosensitivity of NPC cells. In vivo assays extended the significance of these results, showing that miR-124 overexpression decreased cell resistance to radiation treatment in tumor xenografts. Furthermore, we identified PDCD6 as a novel direct target of miR-124. Functional studies showed that knockdown PDCD6 enhanced cell radosensitivity to irradiation, and PDCD6 could rescue the effect caused by overexpression of miR-124, indicating that PDCD6 is a functional target of miR-124. Conclusions: MiR-124 enhances cell radiosensitivity by targeting PDCD6, miR-124/PDCD6 axis may facilitate the development of novel targeted therapies for NPC.
Keywords: miR-124, PDCD6, radiosensitivity, NPC
Introduction
Nasopharyngeal carcinoma (NPC) is a common head and neck malignancy in southern China and southeastern Asia, and radiotherapy is the mainstay of treatment [1,2]. Although advances have been made in the clinical treatment of NPC, patient outcomes have remained unsatisfactory over the last few years mainly due to radioresistance [3,4]. However, the underlying molecular mechanisms of NPC radioresistance remain poorly understood. It is therefore essential to gain a better understanding of the molecular mechanisms underlying NPC radiosensitivity to improve treatment efficacy.
MicroRNAs (miRNAs) are a class of small (~22 nucleotides) noncoding RNA molecules that regulate posttranscriptional gene expression. Accumulating evidence suggests that MiRNAs (miRs) are believed to play fundamental roles in the human cancers, and have a great potential for the diagnosis and treatment of cancer [5]. Regulation of tumor radiosensitivity via miRs-associated mechanisms has attracted much attention in the recent years [3,6-8]. Over the past few years, several miRs have been identified involving in tumor radioresistance, such as miR-23b, miR-95, miR-21, let7 and so on [9-11].
MiR-124 is a brain-enriched miRNA, which is significantly down-regulated in many human malignant tumors, such as gastric carcinoma, glioblastoma, medulloblastoma, colon cancer and hepatocellular carcinoma [12-18]. Recent studies have showed miR-124 could enhance cell radiosensitivity by downregulating the expression of downstream gene in different tumors. However, whether miR-124 has an impact on radiosensitivity in NPC cells remains unknown.
Programmed Cell Death Protein 6(PDCD6), apoptosis-linked gene-2 (ALG-2), is a calcium-binding modulator protein associated with cell proliferation and death. It is originally described as a pro-apoptotic protein in a functional screen of T-cell hybridoma cells [19]. The PDCD6 protein, which is a 22-kDa Ca2+-binding protein containing five serially repetitive EF-hand structures, is one of the prototypic members of the penta EF-hand protein family, participating in T cell receptor, Fas, and glucocorticoid-induced programmed cell death [20,21]. Furthermore, PDCD6 was found to be up-regulated in a variety of tumors compared to normal tissues of the breast, liver, lung, and colon, especially in metastatic tissues, suggesting that in addition to its known pro-apoptotic function PDCD6 may play a role in cell survival [22,23]. In contrast, down-regulation of PDCD6 expression was also observed in gastric cancer and HeLa cells [24,25].
In this study, we proved overexpression of miR-124 or PDCD6 knockdown could enhance NPC cell radiosensitivity, while restoration of PDCD6 in miR-124 transfected cells could rescue the effects. We identified and confirmed PDCD6 as a novel direct target of miR-124 using target prediction algorithms and luciferase reporter gene assays. In summary, we demonstrated miR-124 sensitized NPC cells to radiation treatment at least partially by downregulating PDCD6.
In this study, we found that miR-124 expression was downregulated in both NPC cell lines and tissues; overexpression of miR-124 could increase NPC radioresistance both in vitro and in vivo. PDCD6 was identified as a direct target of miR-124, restoration of PDCD6 in miR-124-overexpresses cells could offset the effect caused by miR-124. Our study for the first time shows the role and mechanism of miR-124 in tumor radioresistance, which highlights the radiosensitizing potential of miR-124/PDCD6 axis in NPC and perhaps in other cancers. Thus, these findings provide valuable clues toward understanding the molecular mechanisms that regulate the pathogenesis of NPC and may be helpful in raising NPC cell radiosensitivity and improve the treatment of NPC in the future.
Materials and methods
Patient specimens and RNA extraction
Primary NPC biopsy specimens and normal biopsies of the nasopharynx were obtained from the First Affiliated Hospital of Jinan University (Guangzhou, China). Both tumor and normal tissues were histologically confirmed by H&E (hematoxylin and eosin) staining. Written informed consents were obtained from all study participants. Collections and using of tissue samples were approved by the Ethics Committee of Jinan University. RNA was extracted from fresh tissues using the AmbionmirVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions.
Cell lines and reagents
An immortalized nasopharyngeal epithelial cell NP69 was cultured in Keratinocyte-SFM (Invitrogen) supplemented with bovine pituitary extract (BD Biosciences). The human NPC cell lines 5-8F, 6-10B, CNE1, CNE2, C666-1, HONE1, and HNE-1 were cultured in RPMI-1640 (Invitrogen). HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen). MiRNAs were transfected at a working concentration of 100 nmol/L using Lipofectamine 2000 reagent (Invitrogen). The miR-124 mimic, a nonspecific miR control, anti-miR-124, and a nonspecific antimiR control were all purchased from Dharmacon. Full-length PDCD6 cDNA entirely lacking the 3’-UTR was purchased from GeneCopeia (Rockville, MD, USA) and subcloned into the eukaryotic expression vector pcDNA3.1(+) (Invitrogen). The pre-miR-124 sequence was amplified and cloned into pCDH-CMV-MCS-EF1-coGFP constructs (System Biosciences, California, USA). Virus particles were harvested 48 h after pCDH-CMV-miR-124 transfection with the packaging plasmid pRSV/pREV, pCMV/pVSVG and pMDLG/pRRE into 293T cells using Lipofectamine 2000 reagent (Invitrogen). PDCD6 knockdown and control lentiviruses were purchased from GENECHEM (Shanghai, China).
RNA isolation, reverse transcription, and quantitative real-time PCR
Total RNA was extracted from cells with TRIzol reagent (Invitrogen). For miR-124, reverse transcription and qRT-PCR reactions were performed by means of a qSYBR-green-containing PCR kit (GenePharma, Shanghai, China), and U6 snRNA was used as an endogenous control. The precursor form of miR-124 was amplified. For detection of PDCD6 mRNA, cDNA was synthesized from 1 µg of total RNA by means of the reverse transcription reaction kit according to the manufacturer’s instructions (Promega). Human GAPDH was amplified in parallel as an internal control. The primers were listed in Supplementary Table 1. All samples were normalized to internal controls and fold changes were calculated through relative quantification (2-ΔΔCt).
Western blot analysis
Equal amounts of protein were resolved by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membranes were blocked in 5% non-fat skim milk/TBST (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.1% Tween-20) at room temperature for 1 h and detected with primary antibodies at 4°C overnight. It was hen blotted for 1 h at room temperature with an appropriate secondary antibody, followed by treatment with enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech, USA). The primary antibodies PRRX1 was purchased from Abcam (UK), Caspase-3, Bcl-2, PDCD6 and γ-H2AX were purchased from Epitomics (USA).
Luciferase assay
The full-length PDCD6 3’-UTR was amplified by PCR and cloned downstream of the firefly luciferase gene in the pGL3 vector (Promega). The vector was named wild-type (wt) 3’-UTR. Site-directed mutagenesis of the miR-124 binding site in PDCD6 3’-UTR was performed using GeneTailor Site-Directed Mutagenesis System (Invitrogen) and named mutant (mt) 3’-UTR. Cells were transfected with reporter plasmids and placed in 96-well plates. After 48 h of incubation, the cells were harvested and assayed using the dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s instructions. Luciferase activities were normalized by β-galactosidase activity. Each experiment was repeated at least three times in triplicate.
Irradiation and clonogenic assay
Cells treated as described were seeded on six-well plates in triplicate and exposed to radiation the doses indicated using a 6-Mvx-ray generated by a linear accelerator (Varian 2300EX at a dose rate of 5 Gy/min. After incubation at 37°C for 14 days, cells were fisxed in 100% methanol and stained with 1% crystal violet. Colonies containing &50 cells were counted under a light microscope. The surviving fraction was calculated as described previously [26]. At least three independent experiments were performed.
Xenograft studies
All of the animal experiments were carried out in strict adherence with the Regulations for the Administration of Affairs Concerning Experimental Animals. All procedures involving animals and their care in this study were approved by the Institutional Animal Care and Use Committee. For xenograft tumor assays, 1×106 cells treated as indicated were injected subcutaneously into the back of 4 to 6-week-old nude mice. When the tumor volume reached 200 mm3, it was irradiated with a single 10 Gy dose 10 days after the injection. Tumor size was calculated every two days using the following formula: (length×width2)/2.
Statistical analysis
All values are expressed as the mean ± standard deviation. Results were analyzed by ANOVA or using a two-tailed Student’s t test with statistical significance established at P<0.05. All Western blot results were quantified using Bio-Rad lab image quantitative analysis software. All data are presented as the mean ± SD of results from three independent experiments.
Results
miR-124 is downregulated in NPC tissues and cell lines
A panel of human NPC cell lines were first analyzed to quantitate the expression level of miR-124. Results showed that miR-124 was decreased in six NPC cell lines examined compared with the immortalized nontumorigenic cell line NP69 (Figure 1A).
Figure 1.
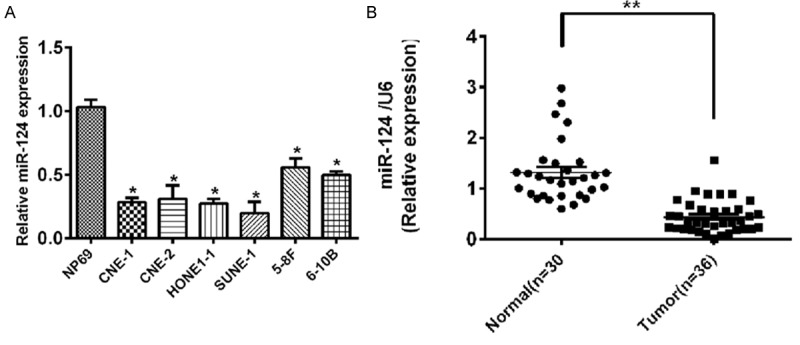
Expression of miR-124 in primary NPC tissues and cell lines. A. miR-124 expression in NPC cell lines compared with nasopharyngeal normal cell line NP69. U6 was used as an internal control. *P<0.05. B. miR-124 expression in NPC tissue compared with matched normal tissue. U6 was used as an internal control.
We further examined miR-124 expression in 36 NPC specimens and 30 normal nasopharyngeal epithelial tissues. Consistent with the data obtained from NPC cell lines, the average expression level of miR-124 was significantly lower in NPC specimens than in normal nasopharyngeal epithelial tissues (Figure 1B; P<0.001).
MiR-124 increases NPC cells sensitivity to radiation treatment
We found overexpression of miR-124 decreased the survival fraction of NPC cells subjected to different doses of irradiation (Figure 2A). In addition, miR-124 decreases activity of Bcl-2, but increases activity of caspase-3 and phosphorylation of the histone H2AX (γ-H2AX) (Figure 2B), an indicator of the cellular response to DNA damage [27].
Figure 2.
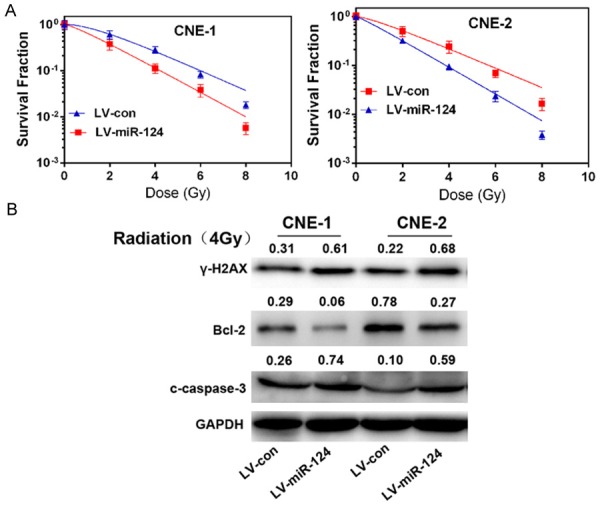
miR-124 enhances NPC cells sensitivity to radiation treatment. A. The survival fraction of CNE-1 and CNE-2 cells treated as described. Colonies containing &50 cells were counted by microscopic inspection, and the surviving fraction was calculated using the multi-target single-hit model: SF=1-(1-e-D/D0)N. The data are presented as the mean ± SD of results from three independent experiments. **P<0.01. B. Western blot analysis of γ-H2AX, Bcl-2, cleaved caspase-3 (c-caspase-3) in CNE-1 and CNE-2 cells continuously overexpressing miR-124 (LV-miR-124), scrambled negative controls (LV-con) treated with 4 Gy dose of radiation. All of these proteins were detected 24 h after radiation, except for γ-H2AX, which was detected 6 h after exposure to radiation.
PDCD6 is a direct target of miR-124
We found a perfect base pairing between the seed sequence of miR-124 and the 3’-UTR of PDCD6 mRNA by using TargetScan (Figure 3A). We then cloned the target sequence of wild-type PDCD6 3’-UTR (wt 3’-UTR) or the mutant sequence (mt 3’-UTR) into a luciferase reporter vector (Figure 3B). HEK293 cells were transfected with a wt or mt 3’-UTR vector together with miR-124 mimics. This resulted in a significant decrease in luciferase activity compared with miRNA controls (Figure 3B, Lanes 2 and 3; P<0.05). The mt 3’-UTR vector was not affected by simultaneous transfection with miR-124 mimics (Figure 3B, Lanes 7 and 8). Furthermore, cotransfection with the anti-miR-124 and wt 3’-UTR vector in HEK293 cells were followed by an increase in luciferase activity (Figure 3B, Lanes 4 and 5; P<0.05). Western blot and qRT-PCR analysis indicated PDCD6 activity had an negative correlation with miR-124 (Figure 3C and 3D).
Figure 3.
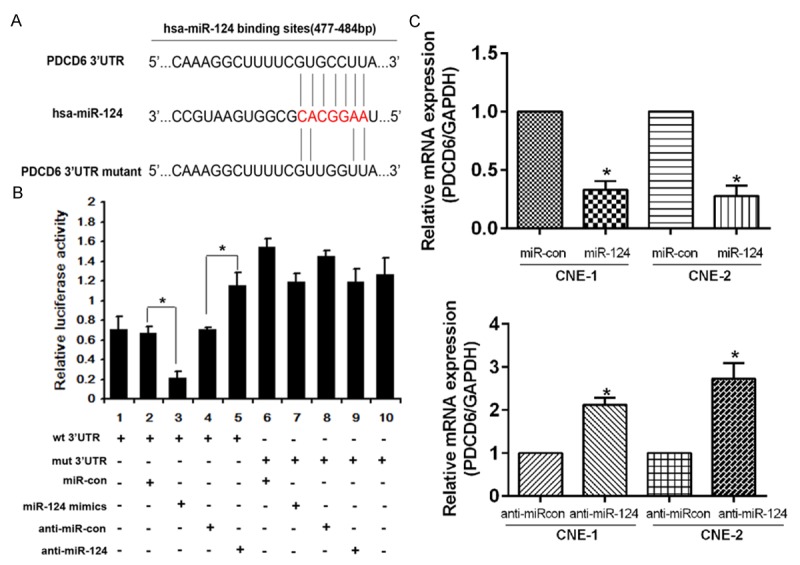
PDCD6 is a direct target of miR-124. A. Wild-type or mutant miR-124 target sequences in the human PDCD6 3’-UTR. B. Luciferase activity assays were carried out using a luciferase reporter with wild-type (wt) or mutant (mt) human PDCD6 3’-UTRs after co-transfection with miR-124 mimics or inhibitor into HEK293 cells. The mt 3’-UTR level increased significantly compared with the wt 3’-UTR. The bar graph shows the mean ± SD of results from three independent transfection experiments. *P<0.05. C. PDCD6 expression in NPC cells treated as described were detected using qRT-PCR. ANOVA and the Student’s t test were used to determine the statistical significance of the differences among groups. *P<0.05.
PDCD6 knockdown confers radiosensitivity in NPC cell lines
Stable cell lines with shRNA-PDCD6 were established, and the PDCD6 protein expression in these cells was verified by western blot analysis of the cell lysates with specific antibodies (Figure 4B). Colony survival assay was subsequently carried out. Clonogenic assay showed PDCD6-silenced cells surviving fractions of the colonies was decreased significantly compared with control group when exposed to different X-ray doses (Figure 4A). Accordingly, Western blot assay showed that when combined with radiation, PDCD6 silencing induced a significantly down-regulation of Bcl-2 level but an up-regulation of caspase-3 expression and γ-H2AX (Figure 4B).
Figure 4.
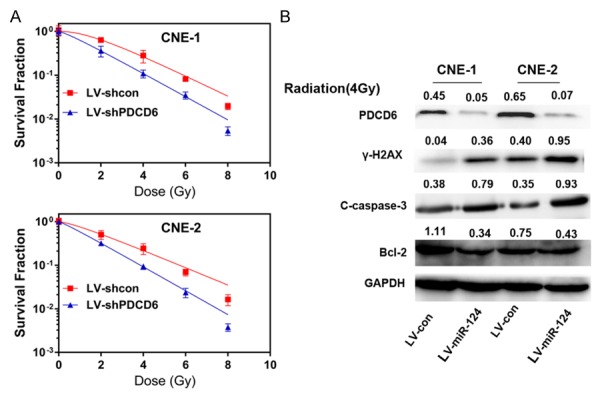
PDCD6 induces cell resistance to radiation in NPC. A. The survival fraction of CNE-1 and CNE-2 cells treated as described was calculated as above indicated. **P<0.01. B. Western blot analysis of γ-H2AX, Bcl-2, cleaved caspase-3 (c-caspase-3) and PDCD6 CNE-1 and CNE-2 cells treated as described. Cells were exposed to a 4 Gy dose of radiation for another 24 h. All of these proteins were detected 24 h after radiation, except γ-H2AX, which was detected 6 h after exposure to radiation.
Enforced expression of PDCD6 restores the effects of miR-124 on NPC cell radiosensitivity
To determine whether the effects of miR-124 were mediated by PDCD6, we transfected pcDNA3.1-PDCD6 into cells overexpressing miR-124. Overexpression of PDCD6 increased the survival fraction of transfected cells compared with non-transfected cells, which suggests that PDCD6 reverses the effects of miR-124 (Figure 5A). Western blot analysis indicated that PDCD6 also restore the expression of caspase-3, γ-H2AX and reduced Bcl-2 expression (Figure 5B).
Figure 5.
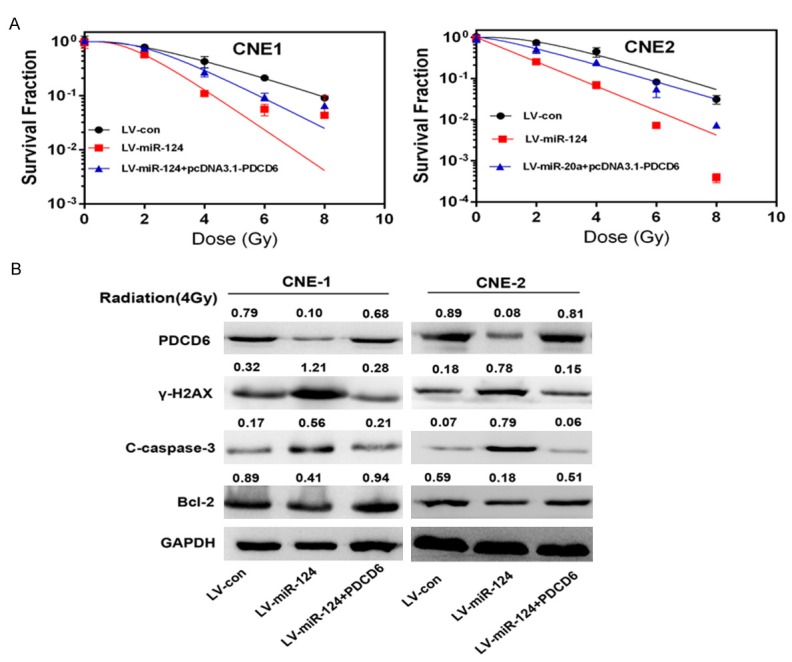
Restoration of PDCD6 in cells overexpressing miR-124 reverses the effects of miR-124 on radiosensitivity. A. The survival fraction of cells treated as described. pcDNA3.1-PDCD6 was transfected into cells continuously overexpressing miR-124 using Lipofectamine 2000. Surviving fraction was calculated as indicated. **P<0.01. B. Western blot analysis of γ-H2AX, Bcl-2, cleaved caspase-3 (c-caspase-3) and PDCD6 expression in CNE-1 and CNE-2 cells treated as described. Cells were exposed to a 4 Gy dose of radiation for another 24 h. All of these proteins were detected 24 h after radiation, except γ-H2AX, which was detected 6 h after exposure to radiation.
miR-124 enhances cell sensitivity to irradiation treatment in vivo
To determine whether miR-124 decreases tumor resistance to the effects of radiation in vivo, we irradiated the tumor tissue using a single 10 Gy dose administered 10 days after injection. We found that the size of the xenografts derived from overexpressing miR-124 was much smaller than the controls exposed to the same dose of radiation (Figure 6A and 6B). These results indicated that miR-124 could sensitize tumors to radiation in vivo.
Figure 6.
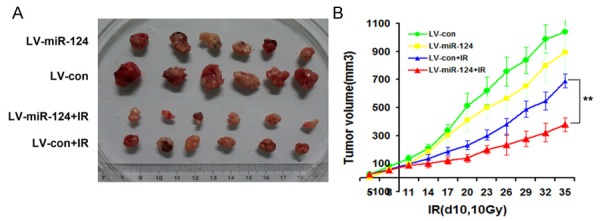
miR-124 enhances cells sensitivity to radiation therapy in vivo. A. Nude mice were subcutaneously injected (into the back of them) with 1×106 cells overexpressing miR-124 or negative control with or without combination of irradiation. B. Overexpression of miR-124 with or without combination of IR in Bel-7402 tumor xenografts by calculating xenografts volumes. Points, mean of tumor volume (mm3) of each treatment group (n=6); bars indicate SE. **P<0.01.
Discussion
miRNAs are known as key regulators of numerous cellular processes, and abnormal expression of miRNAs maybe deemed closely related to the initiation and progression of malignant tumors [28,29]. In this study, we observed that miR-124 is frequently downregulated in NPC cell lines and fresh clinical specimens. Functional analyses revealed that overexpression of miR-124 enhanced the radiosensitivity of NPC cells both in vitro and in vivo. Furthermore, PDCD6 was verified as a direct, functional target of miR-124. Taken together, these data suggest that the identified miR-124/PDCD6 pathway contributes to the elucidation of the mechanisms of radiosensitivity in human NPC and that it may represent a potential target for therapy.
The main problem of treatment failure in many cancers is the resistance of tumor cells to irradiation. Recently, several microRNAs have been reported as playing an important role in regulating therapeutic efficacy in cancer therapy. miR-200c, and miR-199a have been found to induce therapeutic resistance in hepatocellular carcinoma, esophageal, and cervical carcinomas, respectively [30-32]. On the other hand, several microRNAs expressed at lower levels, such as miR-20a, let-7i, miR-181a, miR-630, and miR-128, were identified as closely related to therapeutic resistance in patients with ovarian, lung, and breast cancers, respectively [33-36].
MiR-124 is a brain-enriched miRNA, which is significantly down-regulated in many human malignant tumors, including glioblastoma, gastric carcinoma, medulloblastoma, hepatocellular carcinoma, and CRC [12-18,37,38]. In our recent study, miR-124 was found to be significantly downregulated in NPC tissues and cell lines. To date, the literature remains limited to studies of the biological function and molecular mechanism of miR-124 in NPC. Therefore, we deemed it important to select miR-124 for further investigation in this study.
According to this study, we confirmed firstly that miR-124 was downregulated in NPC cell lines and freshly frozen tissues. We then conducted a series of biological experiments in order to better understand the function of miR-124 on cell radiosensitivityin NPC. Ectopic expression of miR-124 significantly enhanced radiosensitivity both in vitro and in vivo. In a nutshell, these results demonstrate that the enhanced cell radiosensitivity by miR-124 may contribute to the initiation and progression of NPC. For further understanding of the mechanisms underlying, we identified PDCD6 as a potential target gene of miR-124, based on bioinformatics analysis.
Previous studies have elucidated that PDCD6 was up-regulated in a variety of tumors such as breast cancer, liver cancer, lung cancer, and colon cancer, especially in metastatic tissues.Researches suggest that in addition to its known pro-apoptotic function, PDCD6 may play a role in cell survival [22,23]. However, there have been no studies focused on radioresistance of PDCD6 in nasopharyngeal cancer. In this study, a luciferase reporter gene assay verified PDCD6 as a direct target of miR-124. In addition, the overexpression of miR-124 significantly reduced the expression of PDCD6. PDCD6 knockdown could enhance cell radiosensitivity. Moreover, the effect of miR-124 on cell radiosensitivity could be restored by transfecting PDCD6, indicating that PDCD6 is a functional target of miR-124 in NPC.
Conclusion
In this study, we provide evidence that miR-124 can efficiently mediate radioresistance in NPC cell lines by targeting PDCD6, suggesting that miR-124 is an attractive prognostic/predictive biomarker in this type of cancer. Targeting miR-124 may have potential value as an adjuvant therapy for tumor radiosensitization, while more knowledge is required regarding the underlying molecular mechanisms.
Acknowledgements
This work was supported by grants from the Research and Cultivation Fund of Jinan University (No.11616333), Medical Research Fund of Guangdong Province (No.A2016352), The National Natural Science Foundation of China (Grant No.81401973, the Natural Science Foundation of Guangdong Province, China (Grant No.2014A030310040) and Pearl River Nova Program, Science and Technology Program of Guangzhou, China (Grant NO.201710010092).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Feng XP, Yi H, Li MY, Li XH, Yi B, Zhang PF, Li C, Peng F, Tang CE, Li JL, Chen ZC, Xiao ZQ. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 2010;70:3450–3462. doi: 10.1158/0008-5472.CAN-09-4099. [DOI] [PubMed] [Google Scholar]
- 2.Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol. 2012;22:79–86. doi: 10.1016/j.semcancer.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio- and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684:8–18. doi: 10.1016/j.ejphar.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 4.Lee AW, Poon YF, Foo W, Law SC, Cheung FK, Chan DK, Tung SY, Thaw M, Ho JH. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976-1985: overall survival and patterns of failure. Int J Radiat Oncol Biol Phys. 1992;23:261–270. doi: 10.1016/0360-3016(92)90740-9. [DOI] [PubMed] [Google Scholar]
- 5.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhry MA. Radiation-induced microRNA: discovery, functional analysis, and cancer radiotherapy. J Cell Biochem. 2014;115:436–449. doi: 10.1002/jcb.24694. [DOI] [PubMed] [Google Scholar]
- 7.O’Kelly F, Marignol L, Meunier A, Lynch TH, Perry AS, Hollywood D. MicroRNAs as putative mediators of treatment response in prostate cancer. Nat Rev Urol. 2012;9:397–407. doi: 10.1038/nrurol.2012.104. [DOI] [PubMed] [Google Scholar]
- 8.Zhao L, Bode AM, Cao Y, Dong Z. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis. 2012;33:2220–2227. doi: 10.1093/carcin/bgs235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma W, Ma CN, Li XD, Zhang YJ. Examining the effect of gene reduction in miR-95 and enhanced radiosensitivity in non-small cell lung cancer. Cancer Gene Ther. 2016;23:66–71. doi: 10.1038/cgt.2016.2. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, Chen Z, Meng Z. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143. e1112. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Zhang C, Hu L, He Y, Shi Z, Tang S, Chen Y. Abnormal expression of miR-21 and miR-95 in cancer stem-like cells is associated with radioresistance of lung cancer. Cancer Invest. 2015;33:165–171. doi: 10.3109/07357907.2015.1019676. [DOI] [PubMed] [Google Scholar]
- 12.Liu K, Zhao H, Yao H, Lei S, Lei Z, Li T, Qi H. MicroRNA-124 regulates the proliferation of colorectal cancer cells by targeting iASPP. Biomed Res Int. 2013;2013:867537. doi: 10.1155/2013/867537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Lv Z, Yang L. MiR-124 inhibits the growth of glioblastoma through the downregulation of SOS1. Molecular Med Rep. 2013;8:345–349. doi: 10.3892/mmr.2013.1561. [DOI] [PubMed] [Google Scholar]
- 14.Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, Yang Y, McEnery K, Jethwa K, Gjyshi O, Qiao W, Levine NB, Lang FF, Rao G, Fuller GN, Calin GA, Heimberger AB. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913–3926. doi: 10.1158/0008-5472.CAN-12-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, Leung GK, Lu G, Chan DT, Bian XW, Kung HF, Poon WS, Lin MC. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, Xu X, Wu J, Zhu Y, Zheng X, Luo J, Xie L. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013;11:276. doi: 10.1186/1479-5876-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Wang Q, Zhao Q, Di W. MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res. 2013;6:84. doi: 10.1186/1757-2215-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y, Wang K, Wan J. MiR-124 suppresses growth of human colorectal cancer by inhibiting STAT3. PLoS One. 2013;8:e70300. doi: 10.1371/journal.pone.0070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vito P, Lacana E, D’Adamio L. Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science (New York, N.Y.) 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 20.Jung YS, Kim KS, Kim KD, Lim JS, Kim JW, Kim E. Apoptosis-linked gene 2 binds to the death domain of Fas and dissociates from Fas during Fas-mediated apoptosis in Jurkat cells. Biochem Biophys Res Commun. 2001;288:420–426. doi: 10.1006/bbrc.2001.5769. [DOI] [PubMed] [Google Scholar]
- 21.Shibata H, Yamada K, Mizuno T, Yorikawa C, Takahashi H, Satoh H, Kitaura Y, Maki M. The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1. J Biochem. 2004;135:117–128. doi: 10.1093/jb/mvh014. [DOI] [PubMed] [Google Scholar]
- 22.la Cour JM, Mollerup J, Winding P, Tarabykina S, Sehested M, Berchtold MW. Up-regulation of ALG-2 in hepatomas and lung cancer tissue. Am J Pathol. 2003;163:81–89. doi: 10.1016/S0002-9440(10)63632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao RV, Poksay KS, Castro-Obregon S, Schilling B, Row RH, del Rio G, Gibson BW, Ellerby HM, Bredesen DE. Molecular components of a cell death pathway activated by endoplasmic reticulum stress. J Biol Chem. 2004;279:177–187. doi: 10.1074/jbc.M304490200. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Arao T, Gotoda T, Taniguchi H, Oda I, Shirao K, Shimada Y, Hamaguchi T, Kato K, Hamano T, Koizumi F, Tamura T, Saito D, Shimoda T, Saka M, Fukagawa T, Katai H, Sano T, Sasako M, Nishio K. Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples. Cancer Sci. 2008;99:2193–2199. doi: 10.1111/j.1349-7006.2008.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoj BR, la Cour JM, Mollerup J, Berchtold MW. ALG-2 knockdown in HeLa cells results in G2/M cell cycle phase accumulation and cell death. Biochem Biophys Res Commun. 2009;378:145–148. doi: 10.1016/j.bbrc.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Xie G, Zhan J, Tian Y, Liu Y, Chen Z, Ren C, Sun Q, Lian J, Chen L, Ruan J, Ye C, Sun A, Yuan Y. Mammosphere cells from high-passage MCF7 cell line show variable loss of tumorigenicity and radioresistance. Cancer Lett. 2012;316:53–61. doi: 10.1016/j.canlet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 28.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 30.Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, Doki Y. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 31.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zheng L, Ding Y, Li Q, Wang R, Liu T, Sun Q, Yang H, Peng S, Wang W, Chen L. MiR-20a induces cell radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys. 2015;92:1132–1140. doi: 10.1016/j.ijrobp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, Kroemer G. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 34.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber BL, Butzow R, Croce CM, Coukos G, Zhang L. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 37.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silber J, Hashizume R, Felix T, Hariono S, Yu M, Berger MS, Huse JT, VandenBerg SR, James CD, Hodgson JG, Gupta N. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15:83–90. doi: 10.1093/neuonc/nos281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


