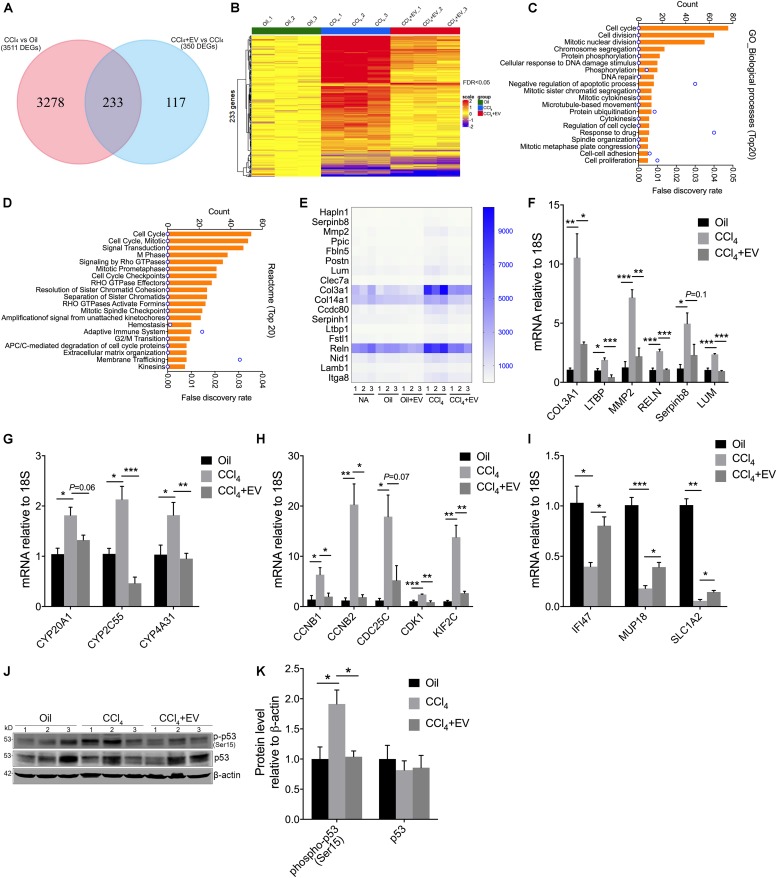
FIGURE 4.
Transcriptomic changes by EVNorm in long-term CCl4-mediated liver injury. Liver tissues from wild type Swiss Webster mice that were control or received 5 weeks of oil or CCl4, with or without EVNorm (80 μg/25 g i.p. qod) over the last 2 weeks, were subjected to transcriptome analysis by RNA-seq as described in Methods. (A) Venn diagram showing DEGs between CCl4 vs. oil and CCl4 + EVNorm vs. CCl4. (B) Heatmap showing pattern of expression of 233 DEGs identified in each animal of each experimental group. (C) Gene ontology (biological processes) analysis of the 233 DEGs showing the top 20 enriched biological processes. (D) Top 20 Reactome pathways of the 233 DEGs. (E) Heat map showing differential expression of interacting ECM-related genes identified by STRING analysis (see Supplementary Figure 2). qRT-PCR showing the effect of EVNorm on either hepatic expression of CCl4-induced (F) fibrosis-related ECM genes, (G) cytochromes, (H) cell cycle-related genes, or on (I) CCl4-suppressed genes. (J) Liver tissue levels of total p53 or phopho-p53 detected by Western blot in light of identification of p53 by Ingenuity Pathway Analysis (Supplementary Figure 3). (K) Quantification of data in panel (J) using ImageJ. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005.