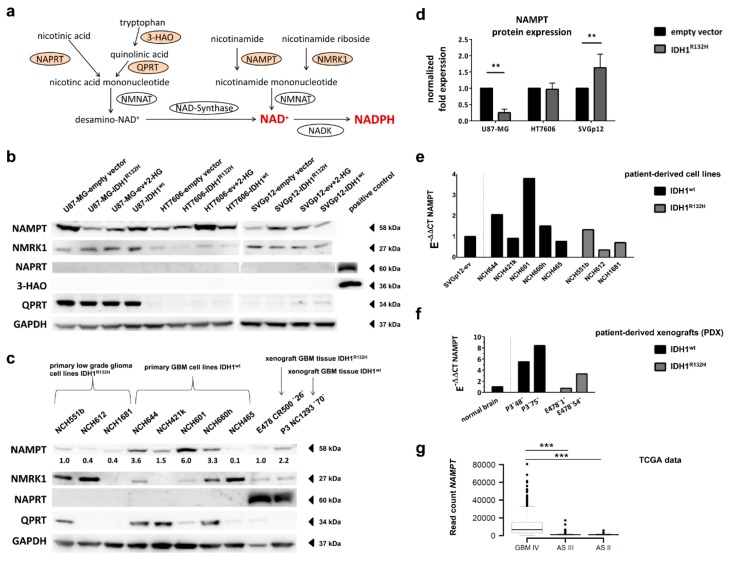
Figure 4.
IDH1-mutation influences the expression of the NAD+ synthesis enzyme NAMPT: (a) NAD synthesis and salvage pathway (adapted from [27]). NAD is synthesized de novo from tryptophan or from nicotinic acid, nicotinamide riboside and nicotinamide via a salvage pathway. NAD-Kinase (NADK) generates NADPH from NAD+ and ATP. NAPRT: nicotinicacid phosphorybosiltransferse, NMRK1: nicotinamide riboside kinase, NAMPT: nicotinaminde phosphoribosyltransferase, QPRT: quinolinic acid phosphoribosyltransferase, 3-HAO: quniolinicacid-synthesis-enzyme 3-hydroxyanthranilate 3,4-dioxygenase. (b) Representative Western Blot showing the expression of NAD-Synthesis enzymes NAMPT, NMRK1, NAPRT, QRPT and 3-HAO in our cell models. GAPDH was applied to determine protein loading. (c) Proteins expression of NAD-Synthesis enzymes in patient-derived cell lines and xenografts (PDX) tissues of IDH1R132H-mutant and IDH-wildtype gliomas. To quantify NAMPT protein levels, density per sample was divided by the loading-control (GAPDH) and relative density for that lane is given. (d) Mean values of NAMPT protein expression normalized to GAPDH and the empty vector controls from four Western Blots performed with protein extracts of different transductions are shown. (e,f) NAMPT RNA expression levels were measured using reverse transcription quantitative PCR (RT-qPCR). Gene expression was normalized to the expression of reference genes GAPDH and ARF1 (E-ΔCT) and thereafter to the expression in astrocytes or commercially available RNA from normal brain control tissue (E-ΔΔCT). (g) Box plots showing gene expression of NAMPT in IDH1-wildtype (GBM IV) and IDH1-mutant (AS III and AS II) glioma patients based on The Cancer Genome Atlas (TCGA) RNA-seq normalized read count data. Box limits indicate 25th and 75th percentiles, whiskers extend at most to 1.5 times the interquartile range of the box, dots represent outliers. NAMPT was significantly higher expressed in GBM IV versus AS II and III (Wilcoxon rank sum test: p = 1.456541 × 10−26 and p = 1.240315 × 10−36, respectively). (** p < 0.01, *** p < 0.001 one-way analysis of variance (ANOVA) followed by Dunnett’s post-hoc t-test).