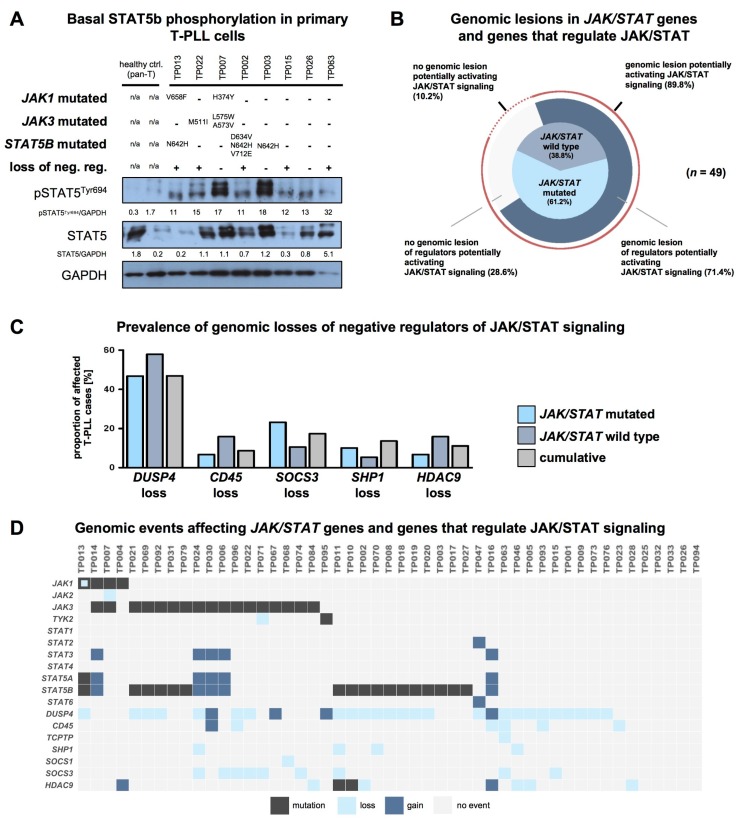
Figure 4.
T-PLL cells show basal STAT5B phosphorylation, regardless of their JAK/STAT mutation status. (A) Elevated basal STAT5B phosphorylation levels of T-PLL cells. Controls: CD3+ pan T-cells isolated from healthy individuals. ‘Loss of neg. reg.’: key regulators which negatively affect STAT5B activation (namely DUSP4, CD45, TCPTP, SHP1, SOCS1, SOCS3, and HDAC; see (D) for case-wise depiction) were considered based on available literature and based on data of their copy number alterations (CNA) in T-PLL. (B) Distribution of genomic lesions affecting any JAK or STAT gene and their regulators (n = 49 cases analyzed with WES/WGS and with SNP arrays). Inner pie chart: distribution of JAK/STAT mutations. Outer pie chart: Prevalence of genomic lesions of regulators activating JAK/STAT (either genomic losses of negative regulators or genomic gains of positive regulators). An overall proportion of 89.8% of T-PLL cases carried a genomic lesion potentially explaining constitutive STAT5B activation (mutation or CNA of JAK/STAT regulator). (C) Prevalence of the five most common genomic lesions affecting negative regulators of the JAK/STAT pathway (n = 49 cases analyzed with WES/WGS and SNP array): T-PLL cases without any mutation in a JAK or STAT gene showed a higher prevalence of genomic losses of DUSP4, CD45, and HDAC9 as compared to JAK/STAT mutated cases. (D) Mapped genomic events involving JAK and STAT genes and their regulators (n = 49 cases analyzed with WES/WGS and SNP array). An overview of genomic lesions resulting in a suggested activation of the JAK/STAT pathway across all considered T-PLL cases is given in Figure S3.