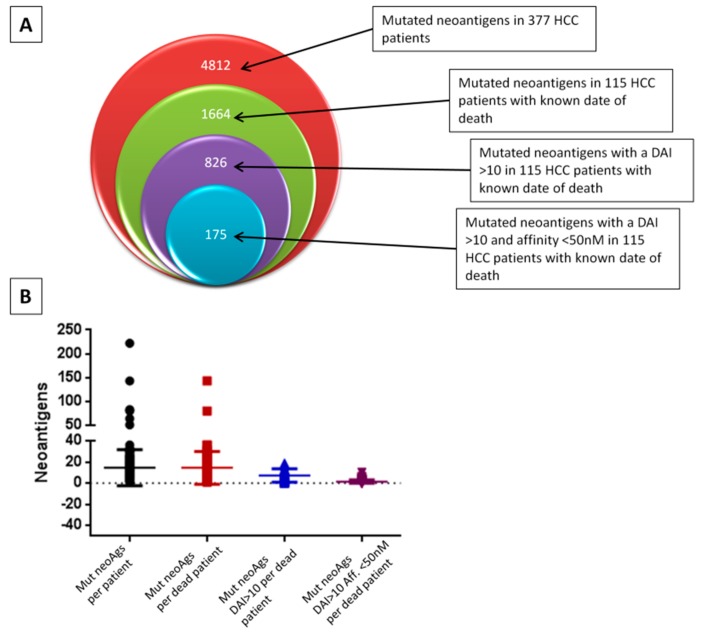
Figure 2.
Prediction in HCC samples. (A) Neoantigens were predicted by NetMHCstabpan algorithm from the nsSNVs identified in the HCC samples at The Cancer Genome Atlas (TCGA) database. (B) Graphic representation of the number of neoantigens predicted in each HCC patient. DAI: differential agretopicity index. nsSNVs: non-synonymous single nucleotide variations.