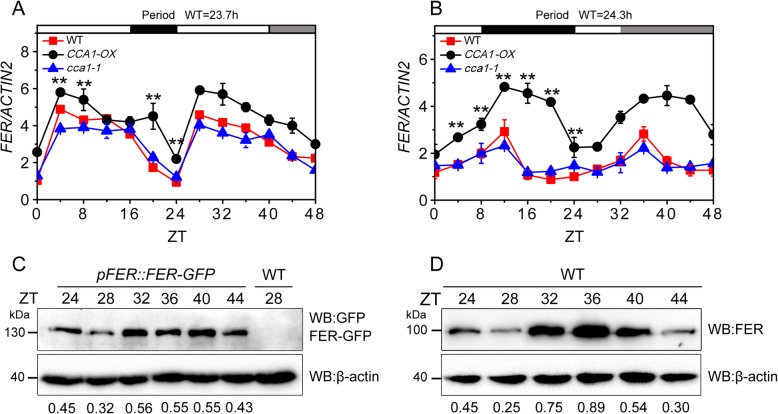
Fig. 1.
FER expression oscillates at a diurnal rhythm. a FER expression in CCA1-OX, cca1–1 and WT plants under LD condition. Seedlings sampled at 4-h intervals were analyzed by qPCR. Day, night, and subjective night are denoted by white, black, and gray bars. ZT 0 represents the light-on time of the day, during which the sample was collected. ACTIN2 was used as an internal control to calculate the relative mRNA levels; the experiments were repeated three times, and the error bars represent the SD of three technical replicates. The period was analyzed using BioDare2. Asterisks indicate a significant difference between CCA1-OX and WT (**P < 0.01, one-way ANOVA with Tukey’s test). b FER expression in CCA1-OX, cca1–1 and WT plants under SD condition. The experiments were repeated three times, and the error bars represent the SD of three technical replicates. The period was analyzed using BioDare2. Asterisks indicate a significant difference between CCA1-OX and WT (**P < 0.01, one-way ANOVA with Tukey’s test). c Immunoblot analysis. The samples were collected at 4-h intervals. All proteins were extracted from pFER::FER-GFP and WT seedlings and then analyzed using anti-GFP antibodies to detect the FER-GFP protein. The FER-GFP/β-actin ratio is displayed below the gel, and β-actin was used as the loading control. The experiments were independently repeated three times with similar results. d Immunoblot analysis. Protein was extracted from WT seedlings and then analyzed using anti-FER antibodies to detect the FER protein. β-actin was used as the loading control, and the FER/β-actin ratio is displayed below the gel. At least three biological replicates were performed, and similar results were obtained.