Figure 1.
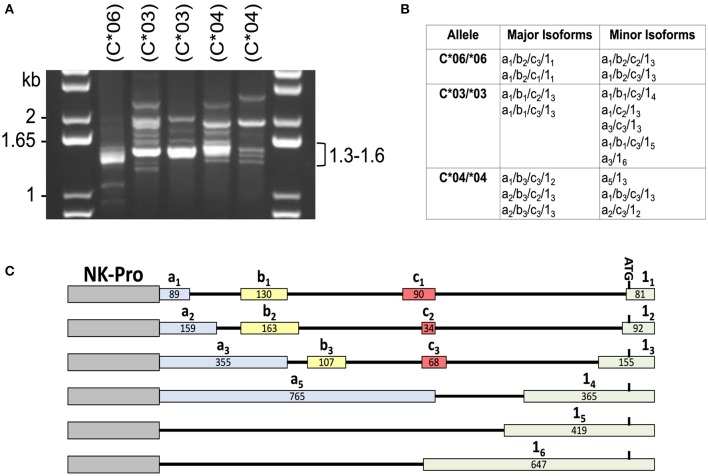
Individuals with homozygous HLA-C alleles exhibit distinct patterns of alternative transcripts. (A) PCR amplification of cDNA from individuals homozygous for specific HLA-C alleles. NK cell cDNA from HLA-C homozygous donors (C*06/*06; C*03/*03; C*04/*04) was amplified with exon -1a forward and exon 8 reverse primers. The size of bands in the DNA marker lanes (kb) and the size range of major bands amplified from the cDNAs is indicated. HLA-C allele is indicated as (C*##) above each lane. (B) Sequencing analysis of PCR products. A summary of the exon structure of translatable mRNA isoforms detected is shown. All isoforms listed contain exons 1–8 comprising the complete open reading frame. The alternative 5′-UTR exons a, b, c, and 1 are numbered as in Li et al. (12). (C) Summary of the HLA-C alternative exons observed in this study. The relative genomic position, name, and size in bp is shown for each of the exons present in the NK-Pro transcripts listed in (B). The locations of the NK-Pro and the ATG initiation codon of the HLA-C gene are indicated.