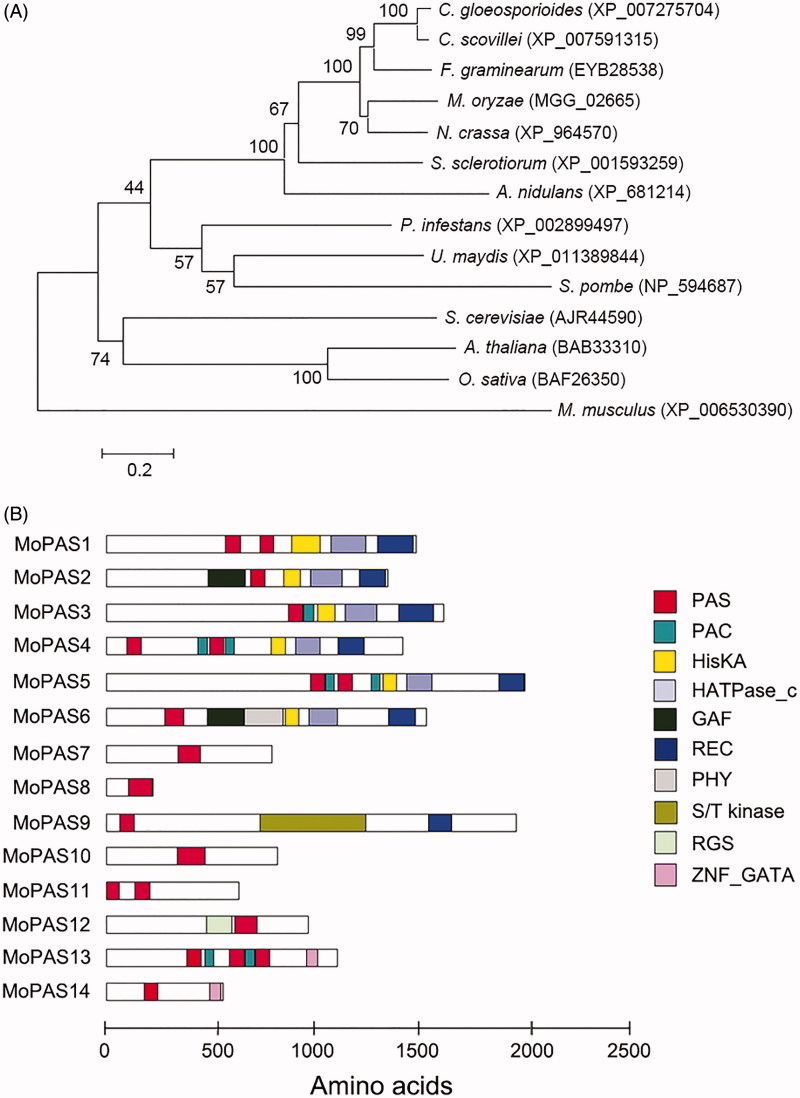
Figure 1.
Phylogenetic analysis of MoPAS1. (A) Phylogenetic tree analysis of MoPAS1. A neighbor-joining tree of the amino acid sequence of PAS proteins. Numbers at nodes represent bootstrap values calculated from 1000 replicates. The scale bar indicates the number of amino acid differences per site. Sequence information was taken from the NCBI protein database. GenBank accession number for each protein is followed by species; (B) Predicted domain structure of MoPAS proteins. Sequence information was taken from the Magnaporthe genome database of the Broad Institute, and domain structure analysis was performed using InterPro Scan (http://www.ebi.ac.uk/interpro/). PAS (period circadian protein, aryl hydrocarbon receptor nuclear translocator protein, single-minded protein; IPR0000014), PAC (C-terminal to PAS motifs; IPR001610), HisKA (signal transduction histidine kinase; IPR003661), HATPase_C (histidine kinase-like ATPase; IPR003594), GAF (domain present phytochrome and cGMP-specific phosphodiesterases; IPR003018), REC (signal receiver domain; IPR001789), PHY (phytochrome; IPR001294), S/T kinase (serine/threonine kinase; IPR008271), RGS (regulator of G protein signaling; IPR016137), ZNF_GATA (zinc finger, GATA-type; IPR000679).