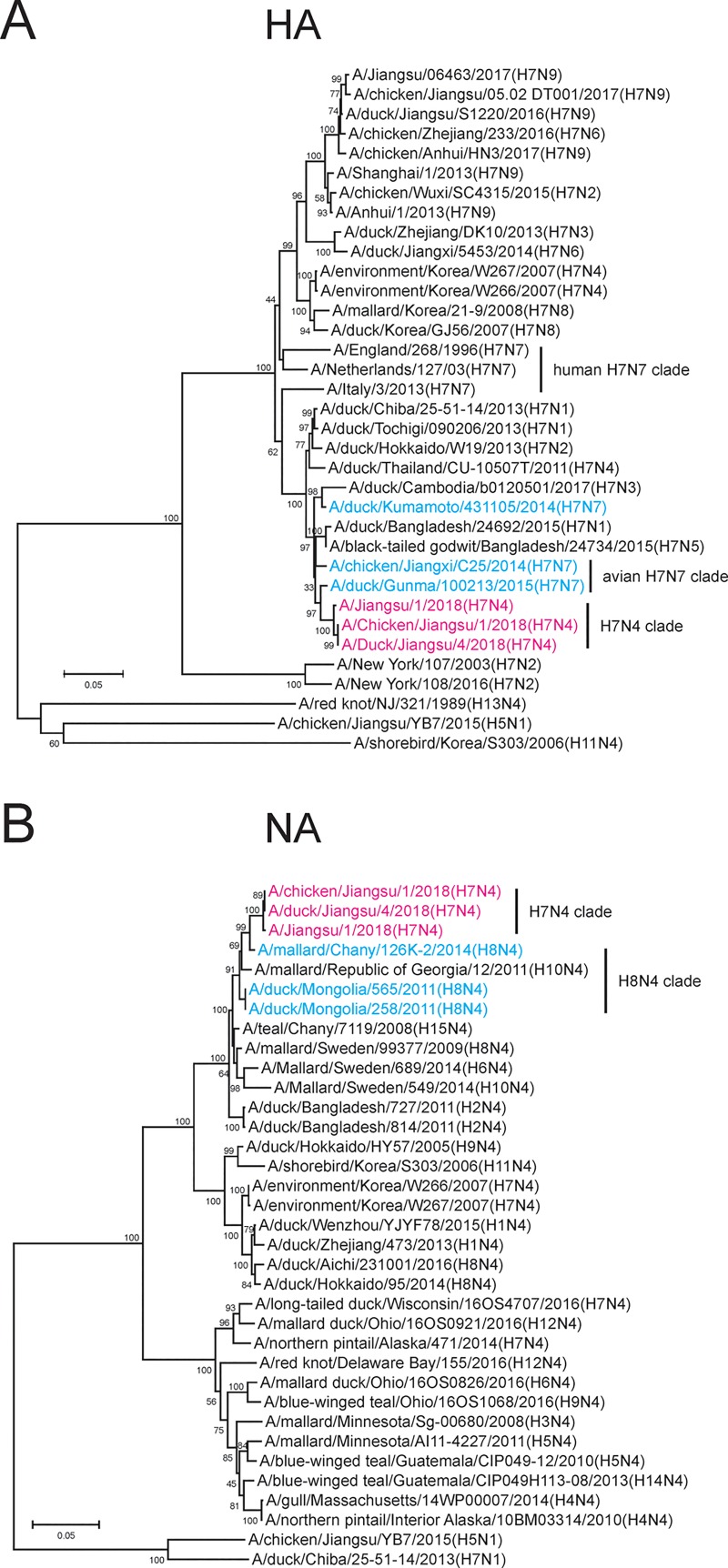
Fig 1. Phylogenetic tree analysis of the origins of HA and NA segments of the H7N4 human isolate in China.
A) 35 HA segments of H7 isolates were analyzed using MEGA6 software. Magenta: new H7N4 viruses including A/Jiangsu/1/2018 (JS2018); Cyan: related avian H7N7 viruses; Black: H7N1, H7N2, H7N3, H7N5, H7N6, H7N7 (human), H7N8 and H7N9 viruses. A/red not/NJ/321/1989 (H13N4), A/shorebird/Korea/S303/2006 (H11N4) and A/chicken/Jiangsu/YB7/2015 (H5N1) isolated in Jiangsu were used as outgroup control strains. Scale bar indicates the amount of substitutions in the nucleotides. B) 35 NA segments of N4 isolates were analyzed using MEGA6. Magenta: H7N4 viruses including JS2018; Cyan: related avian H8N4 viruses; Black: H1N4, H2N4, H3N4, H4N4, H5N4, H6N4, H8N4, H9N4, H10N4, H11N4, H12N4, H14N4, and H15N4 viruses. Outgroup controls: A/chicken/Jiangsu/YB7/2015 (H5N1) and A/duck/Chiba/25-51-14/2013 (H7N1). Scale bar: the amount of nucleotide substitutions.