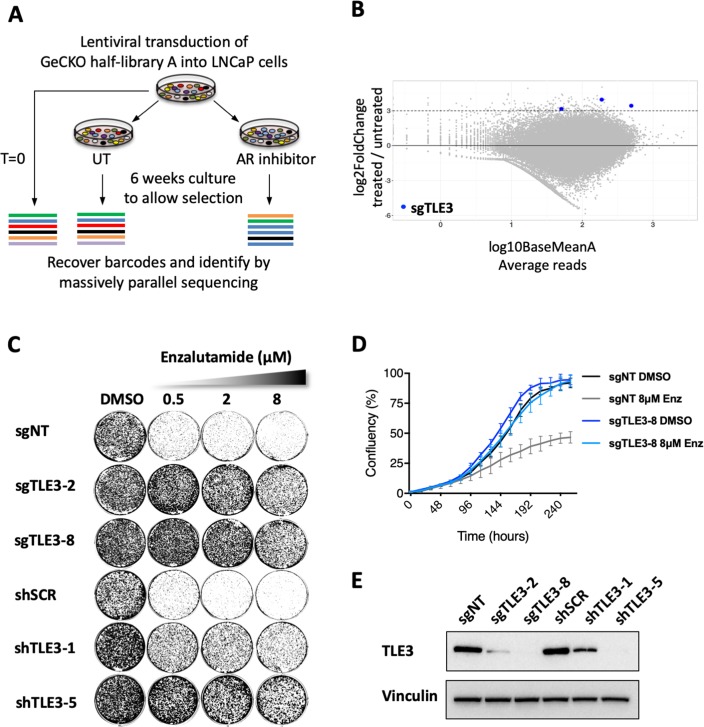
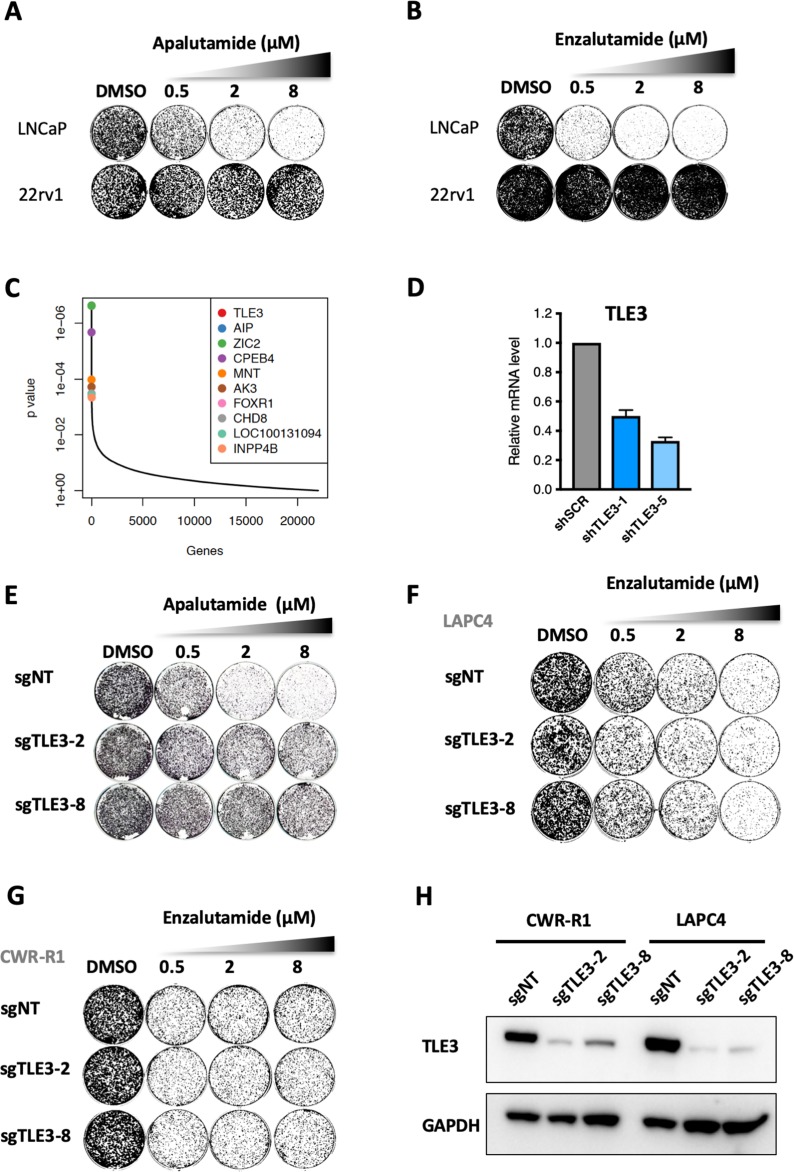
Figure 1. Genome-wide screen identifies TLE3 as a modulator of AR inhibitor sensitivity.
(A) Overview of the genome-wide CRISPR-Cas9 resistance screen. (B) Representation of the relative abundance of the gRNA barcode sequences of the CRISPR-Cas9 resistance screen. The y-axis shows the enrichment (relative abundance of apalutamide treated/untreated) and the x-axis shows the average sequence reads of the untreated samples. (C) Long-term growth assay (14 days) showing the functional phenotype of LNCaP cells harboring TLE3 knockout or knockdown vectors, cultured in the presence of vehicle or enzalutamide. Cells harboring a non-targeting sgRNA (sgNT) or scrambled shRNA (shSCR) were used as a control. (D) Quantitative analysis of live cell proliferation in real-time for control cells and TLE3KO cells in the absence or presence of enzalutamide. (E) Western blot showing TLE3 protein levels for control cells and TLE3KO cells shown in C and D. Vinculin was used as a loading control.