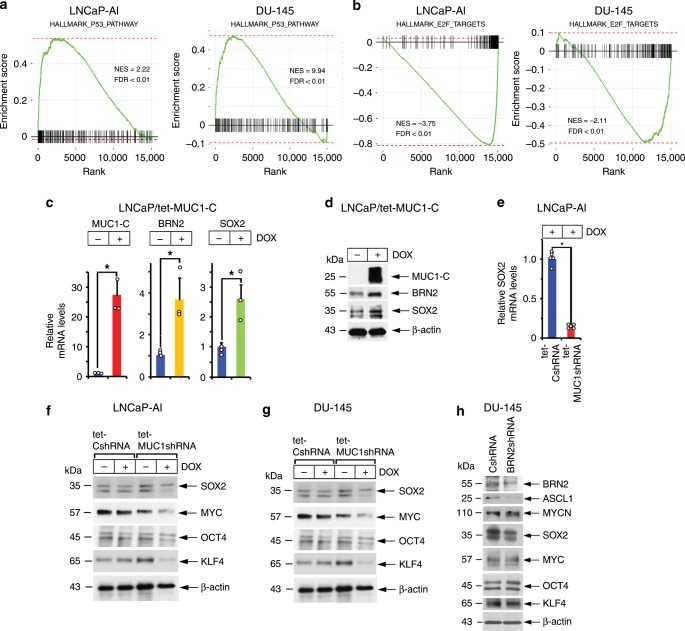
Fig. 5. MUC1-C drives effectors of lineage plasticity.
a RNA-seq was performed in triplicate on LNCaP-AI/tet-MUC1shRNA (left) and DU-145/tet-MUC1shRNA (right) cells treated with vehicle or 500 ng/ml DOX for 7 days. The datasets were analyzed with GSEA, using the Hallmark gene signature collection for the p53 Pathway. b RNA-seq was performed in triplicate on LNCaP-AI/tet-MUC1shRNA (left) and DU-145/tet-MUC1shRNA (right) cells treated with vehicle or 500 ng/ml DOX for 7 days. The datasets were analyzed with GSEA, using the Hallmark gene signature collection for E2F Targets. c and d. LNCaP cells expressing tet-MUC1-C were treated with vehicle or 500 ng/ml DOX for 7 days. MUC1-C, BRN2 and SOX2 mRNA levels were analyzed by qRT-PCR (c). The results (mean±SD of three determinations) are expressed as relative mRNA levels compared to that obtained for vehicle-treated cells (assigned a value of 1). Lysates were immunoblotted with antibodies against the indicated proteins (d). e and f LNCaP-AI cells stably expressing a tet-CshRNA or tet-MUC1shRNA were treated with vehicle or 500 ng/ml DOX for 7 days. SOX2 mRNA levels were analyzed by qRT-PCR (e). The results (mean±SD of five determinations) are expressed as relative mRNA levels compared to that obtained for DOX-treated LNCaP-AI/tet-CshRNA cells (assigned a value of 1). Lysates were immunoblotted with the indicated antibodies (f). g Lysates from DU-145/tet-CshRNA and DU-145/tet-MUC1shRNA cells treated with vehicle or 500 ng/ml DOX for 7 days were immunoblotted with antibodies against the indicated proteins. h Lysates from DU-145/CshRNA and DU-145/BRN2shRNA cells were immunoblotted with antibodies against the indicated proteins. *p < 0.05 (Student’s t-test). Source data are provided as a Source Data file.