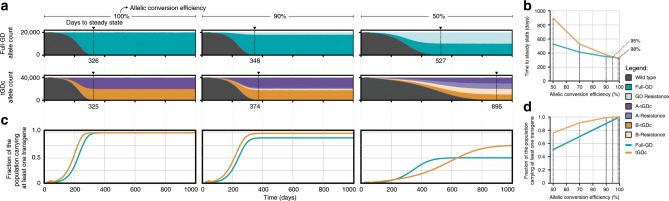
Fig. 6. Comparison of full-drive and tGD spread in modeled populations.
Model predictions for releases of Ae. aegypti mosquitoes homozygous for the tGD and full-drive systems, parameterized with ballpark estimates: (i) a cleavage frequency of 100% in females and males, (ii) an allelic conversion efficiency, given cleavage, of 50–100% in females and males, and (iii) no fitness costs associated with the Cas9 or gRNA alleles. All resistant alleles are assumed to be in-frame/cost-free. Five weekly releases are simulated, consisting of 100 adult males homozygous for each system, into a population having an equilibrium size of 10,000 adults. Model predictions were computed using 50 realizations of the stochastic implementation of the MGDrivE simulation framework. a Stacked allele counts over time for the full-drive and tGD systems for allelic conversion efficiencies of 100, 90, and 50%. b Allelic conversion efficiency plotted against time to steady state for the full-drive (turquoise) and tGD (yellow) systems. c Fraction of the population carrying at least one transgene over time for the full-drive (turquoise) and tGD (yellow) systems for allelic conversion efficiencies of 100, 90, and 50%. d Allelic conversion efficiency plotted against the fraction of the population carrying at least one transgene at equilibrium for the full-drive (turquoise) and tGD (yellow) systems.