Figure 1.
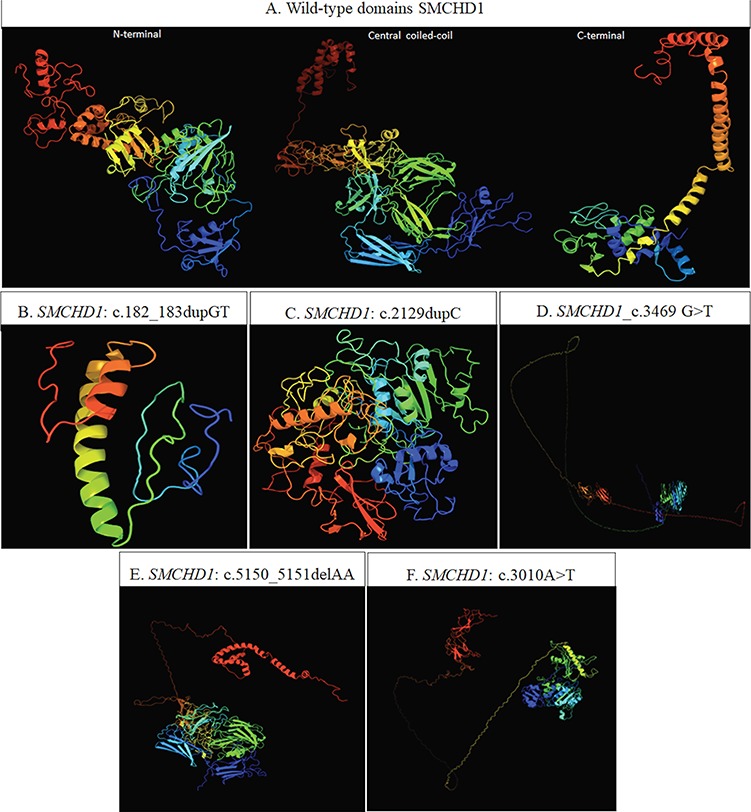
(A) Predicted conformation of the three wild-type domains of SMCHD1, based on the domain organization released by UniProt (entry: A6NHR9). In particular, the N-terminal region (1–702 AA) harboring the GHKL-ATPase domain (111–702 AA) is based on the template c5ix1A (PDB header: transcription; PDB molecule: MORC family CW-type zinc finger protein 3; PDBTitle: crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide). The central coiled-coil domain (703–1719 AA) is based on the template c4e9lA (PDB header: cell adhesion; PDB molecule: attaching and effacing protein, pathogenesis factor; PDBTitle: FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections). The C-terminal region (1720–2005 AA) harboring the SMC hinge domain (1720-1847AA) is based on c2wd5A (PDB header: cell cycle chain: A: PDB molecule: structural maintenance of chromosomes protein 1a; PDBTitle: SMC hinge heterodimer (mouse). (B–F) 3D model predicted by Phyre2 tool. The structure resulting from the presence of c.182_183dupGT (B) is based on the template d1e9ya1 (fold: beta-clip superfamily: urease, beta-subunit). The structures resulting from the presence of the c.2129dupC and c.3469G>T (C and D, respectively) are based on the template c5ix1A (PDB header: transcription. PDB molecule: MORC family CW-type zinc finger protein 3). The structure resulting from c.5150_5151delAA (E) is based on the template c4e9IA (PDB header: cell adhesion; PDB molecule: attaching and effacing protein, pathogenesis factor). The structure referred to the c.3010A>T (F) is based on the template c5ix1A (PDB header: transcription. PDB Molecule: MORC family CW-type zinc finger protein). The 3D model simulation of the SMCHD1_c.1131+2_1131+5delTAAG is not available because the amino acid sequence alteration following this variant cannot be predicted.
