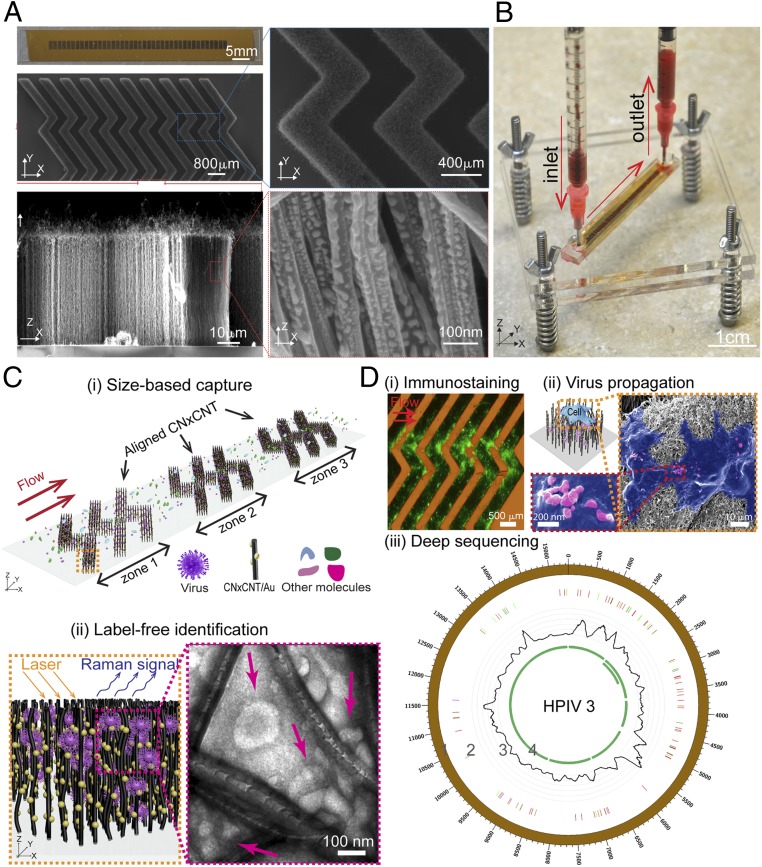
Fig. 1.
Design and working principle of VIRRION for effective virus capture and identification. (A) Photograph and SEM images of aligned CNTs exhibiting herringbone patterns decorated with gold nanoparticles. (B) Picture showing assembled VIRRION device, processing a blood sample. (C) Illustration of (i) size-based capture and (ii) in situ Raman spectroscopy for label-free optical virus identification. Images of electron microscopy showing captured avian influenza virus H5N2 by CNxCNT arrays. (D) On-chip virus analysis and enrichment for NGS, (i) on-chip immunostaining for captured H5N2, (ii) on-chip viral propagation through cell culture, and (iii) genomic sequencing and analysis of human parainfluenza virus type 3 (HPIV 3). Track 1: scale of the base pair position; track 2: variant analysis by mapping to strain #MF973163; color code: deletion (black), transition (A–G, fluorescent green; G–A, dark green; C–T, dark red; T–C, light red), transversion (A–C, brown; C–A, purple; A–T, dark blue; T–A, fluorescent blue; G–T, dark orange; T–G, violet; C–G, yellow; G–C, light violet); track 3: coverage; track 4: regions of open reading frames (ORFs).