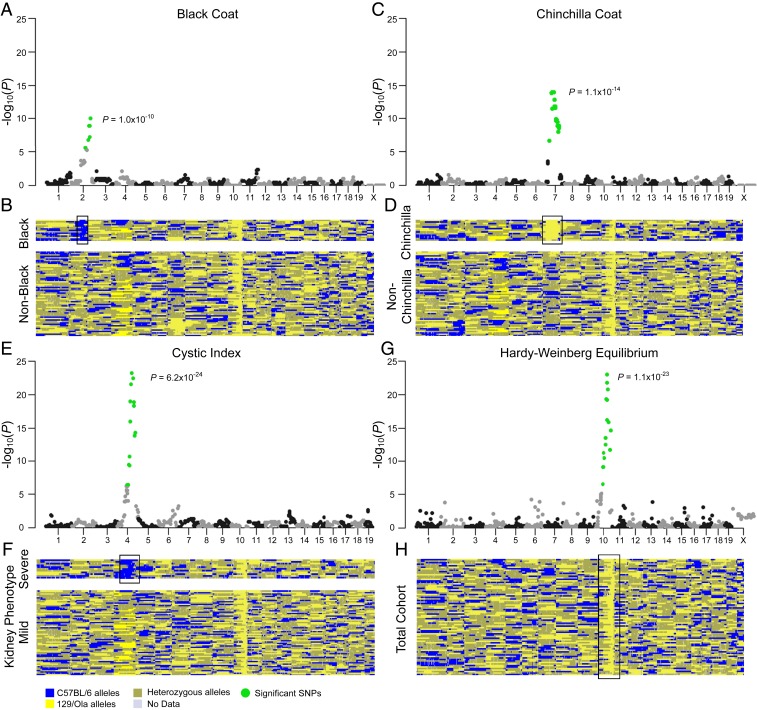
Fig. 2.
Modifier locus in chromosome 4 affects the kidney phenotype in F2 Cep290Gt/Gt mice. (A, C, and E) Manhattan plots showing the strength of association of each variant with coat color (A and C) or severe kidney disease (E) ordered according to genomic position. Significant SNPs are shown in green (P < 6.34 × 10−5). (B, D, and F) Genotyping heatmaps showing alleles inherited from either the C57BL/6 strain (blue) or the 129/Ola strain (yellow). Heterozygous calls are shown in gold. Reads lacking data are shown in gray. Samples are shown Top to Bottom in order of coat color (B and D) or cystic index (F). Mice displaying the phenotype of interest for each plot are shown (Upper) (B, black coat; D, chinchilla coat; F, severe kidney phenotype, cystic index > 10%). (G) Manhattan plot showing the deviation from the Hardy–Weinberg equilibrium at each locus ordered according to genomic position. The only significant SNPs are located adjacent to the locus of Cep290. (H) Genotyping heatmap showing alleles inherited from either parent strain. All mice are shown in a randomized order. The position of significant SNPs is identified in each heatmap (black bounding box). For each plot, the P value of the most significant SNP is shown.