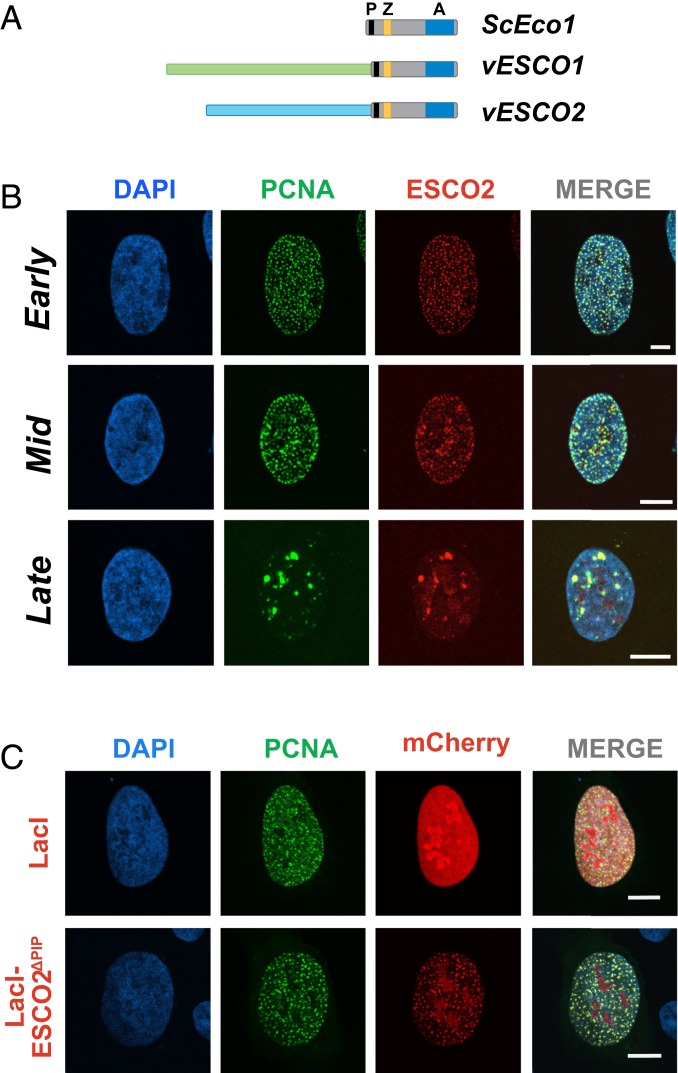
Fig. 1.
ESCO2 associates with sites of active DNA replication. (A) The Eco acetyltransferases. Shown are the S. cerevisiae Eco1 protein and the vertebrate homologs ESCO1 and ESCO2. All 3 proteins contain a highly conserved domain containing a PIP box (P; black), a zinc finger motif (Z; yellow), and the catalytic acetyltransferase domain (A; blue). The vertebrate proteins ESCO1 and ESCO2 have unique N-terminal extensions, with no apparent sequence homology to each other (light green and light blue). (B) ESCO2 colocalizes with PCNA in replication foci. Confocal micrographs of nuclei of U2OS cells cotransfected with mCherry-ESCO2 and GFP-PCNA. ESCO2 colocalized with PCNA in replication foci in patterns of early, mid, and late DNA replication. Sites of colocalization appear yellow in the merged images. (C) The PIP box in ESCO2 is dispensable for localization to replication foci. Confocal micrographs of U2OS cells cotransfected with GFP-PCNA and mCherry-ESCO2 in which the PIP box is deleted. Pearson’s correlation coefficient for red and green signal intensities R = 0.837, 0.754, and 0.591 for early, mid, and late images, respectively (B). R = 0.189 for lacI, and 0.837 for ΔPIP (C). Shown is a representative example of 3 independent experiments. (Scale bars, 10 μm.)