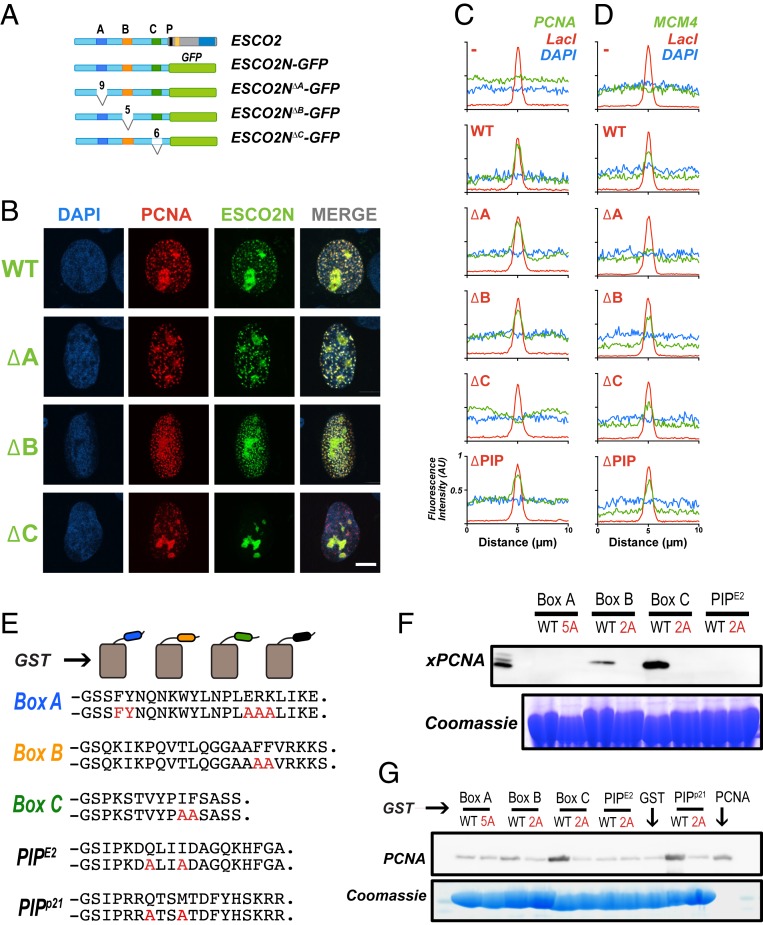
Fig. 4.
PCNA interacts with ESCO2 motifs in vivo and in vitro. (A) ESCO2N-GFP fusions. A cartoon depicting constructs in which the N-terminal 375 amino acids (a.a.) of ESCO2 are fused directly to eGFP. Motifs A, B, and C were deleted independently, as shown; numbers indicate the number of amino acids deleted (not drawn to scale). (B) Localization to replication foci. Confocal images of U2OS cells cotransfected with Ruby-PCNA and the GFP-fusion constructs. Colocalization is indicated in yellow in the merge image, as in Fig. 2. (C) Box C is critical for PCNA recruitment by tethered ESCO2. mCherry-lacI-ESCO2 (full-length) fusions with the deletions indicated in A were coexpressed with GFP-tagged PCNA as in Fig. 2, and the colocalization at nuclear foci was scored by fluorescence intensity profile analysis as in Fig. 2. (D) MCM4 recruitment to tethered ESCO2 is dependent upon box A. The experiment in C was repeated, only in this case the ESCO2 constructs were coexpressed with mEmerald-MCM4. Recruitment to tethered ESCO2 was scored as in Fig. 2. (E) Pull-down assay using GST-fusion proteins. Short peptide sequences including box A, box B, box C, or the ESCO2 PIP box motifs were expressed as GST-fusion proteins. A parallel set was made in which alanine substitutions were made at the invariant amino acids (shown in red). The PIP box from p21 was used as a positive control. (F) Coprecipitation from cell-free extracts. GST-fusion proteins shown in A were mixed with Xenopus egg extract and incubated with glutathione sepharose beads. The beads were washed and bound proteins were eluted and probed for PCNA by immunoblot. A duplicate gel was stained with Coomassie dye to detect the GST-fusion proteins. (G) Coprecipitation of purified proteins. The indicated GST-fusion proteins (E) were mixed with purified recombinant PCNA, pulled down with glutathione agarose beads, and analyzed as in F for PCNA.