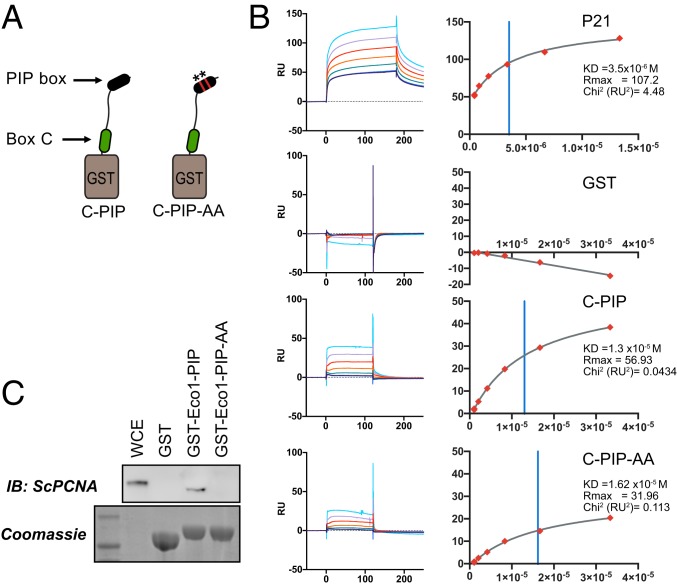
Fig. 5.
Contributions of the canonical PIP motif to PCNA binding. (A) Generation of combined fusions. GST was fused to a peptide spanning amino acids 320 to 388 of human ESCO2 containing both box C and the downstream canonical PIP-like motif at amino acids 374 to 377. A derivative in which the PIP-like motif was mutated (ESCO2 Q374A, I377A) was also purified. (B) Surface plasmon resonance analysis. The GST-fusion peptides in A were used to test PCNA binding using SPR, in which GST fusions were bound by anti-GST antibody and recombinant trimeric PCNA was used as an analyte. Shown are the background-subtracted sensorgrams (Left) and binding curves (Right) for p21 (positive control), GST (negative control), and the indicated fusions. Binding constants are shown at Right. The χ2 residuals are represented as RU2, an indication of curve fitting. Vertical blue lines indicate KD. (C) Contribution of the Eco1 PIP box to PCNA interaction in S. cerevisiae. GST was fused to the first 33 amino acids of S. cerevisiae Eco1p (GST-Eco1-PIP) for use in a pull-down assay (as in Fig. 4). Disruption of the PIP motif by 2 amino acid substitutions (Q18A, L21A = GST-Eco1-PIP-AA) disrupted the ability to pull down PCNA from whole cell yeast extract, as indicated by immunoblot analysis. A parallel gel was Coomassie stained, showing the bead-associated GST fusions. WCE, whole cell yeast extract.