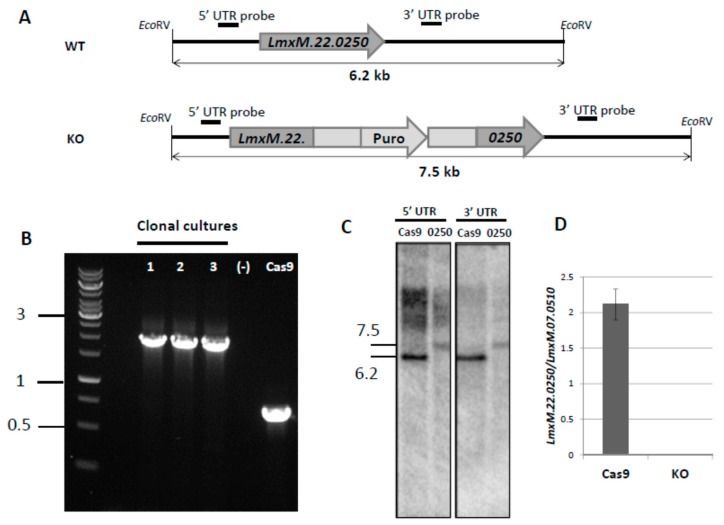
Figure 2.
Knock-out of LmxM.22.0250 with CRISPR-Cas9 approach. (A) Schematic depictions of the WT (Cas9) and recombined alleles after replacement with puromycin-resistant gene; annealing positions of the probes and expected fragment sizes are shown. (B) PCR analysis of LmDUSP1 KO clonal cultures, a wild type (Cas9), and a negative control; 1 kb DNA ladder is on the left. (C) Southern blotting results of the EcoRV digested L. mexicana genomic DNA of the wild type (Cas9, labelled Cas9) and LmxM.22.0250 ablated strains (clonal culture 1 from 2B, labelled 0250) with LmxM.22.0250 5’ UTR and LmxM.22.0250 3’ UTR probes. (D) Quantitative RT-PCR analysis of LmxM.22.0250 gene expression in the wild type (Cas9) and LmDUSP1 KO L. mexicana. Gene expression was normalized to LmxM.07.0510 [29] and presented as means and standard deviations of three independent biological replicates. Sizes in (B,C) are in kb.