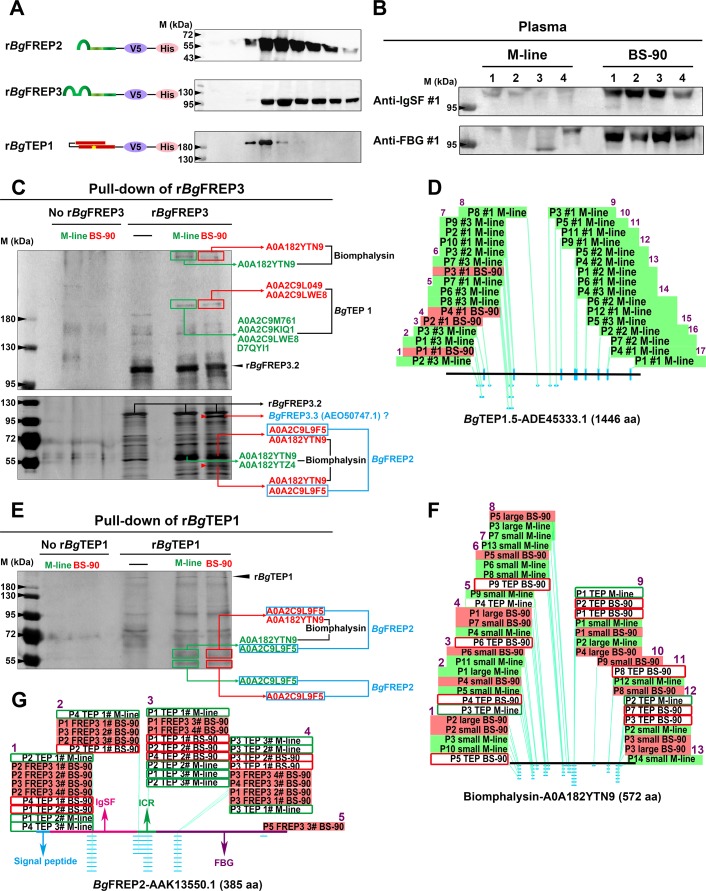
Figure 1. BgFREP3, which had special high abundance in BS-90 plasma, interacted with BgTEP1 and Biomphalysin derived from both M-line and BS-90 plasma, while uniquely associated with BgFREP2 and a BgFREP3 variant from BS-90 plasma.
(A) Schematic diagram and Western blot analysis of recombinant proteins, the ends of which are coupled to the tags V5 and ploy-His for protein immunoassay and purification operations. The yellow star in the schematic structure of rBgTEP1 represents the thioester motif. (B) The background expression of BgFREP3 in BS-90 snail plasma was significantly higher than that in M-line plasma. The haemolymph of four snails from each M-line and BS-90 strain was independently extracted, and haemocytes were removed by centrifugation to obtain plasma samples. After the plasma of each snail was quantified, the same amount of plasma was used for SDS-PAGE electrophoresis and Western blot analysis. Anti-IgSF #1 and anti-FBG #1 antibodies of BgFREP3 were used as primary antibodies. (C) The rBgFREP3 associated with BgTEP1 and Biomphalysin in both M-line and BS-90 plasma, but exclusively interacted with a BgFREP3 variant and BgFREP2 in BS-90 plasma. The eluent of rBgFREP3 pull-downs was collected and separated on 6% (upper panel) and 10% (lower panel) SDS-PAGE gels before silver staining. (E) The rBgTEP1 interacted with Biomphalysin and BgFREP2 in both M-line and BS-90 plasma. The eluent of pull-down experiments with rBgTEP1was separated on 10% SDS-PAGE gels. In the pull-down experiments with rBgFREP3 (C) and rBgTEP1 (E), cell-free plasma from M-line (green) or BS-90 (red) B. glabrata snails was incubated with Sf9 cell lysates expressing rBgFREP3 or concentrated medium containing rBgTEP1 together (interactome experiments, last two lanes) or alone (controls, first three lanes). Bands that differ between control and interactome experiments were cut, proteins submitted to tryptic digest and analyzed by LC-MS/MS for identification. In interactome experiments, bands that differ between M-line and BS-90 lanes were marked with red arrows. The results of mass spectrometry analysis are shown in the figure. Proteins (BgTEP1 and Biomphalysin) represented by black font interacted with rBgFREP3 in both M-line and BS-90 plasma. Proteins (BgFREP2 and BgFREP3.3) represented by blue font specially interacted with rBgFREP3 only in BS-90. (D, F and G) Distribution of peptides identified by LC-MS/MS on BgTEP1.5 (ADE45333.1), Biomphalysin (A0A182YTN9) and BgFREP2 (AAK13550.1). The text on the panels describes the peptides identified from different Pull-down experiments, and most peptides were identified multiple times. The text with the green background describes peptides identified from the plasma of M-line strain in the rBgFREP3 pull-down experiments, and corresponding the text with the red background represents the peptides from the plasma of BS-90 strain. Similarly, the text in the green frame box represents the peptides from the plasma of the M-line strain in the rBgTEP1 pull-down experiments, and corresponding the text in the red frame box represents the peptides from the plasma of the BS-90 strain. The horizontal lines in panels D, F and G represent the full-length amino acid sequences of BgTEP1.5, Biomphalysin and BgFREP2. The light blue dots and dashes indicate the location of the peptides. The purple Arabic numerals represent the number of identified peptide fragments that differ from each other.