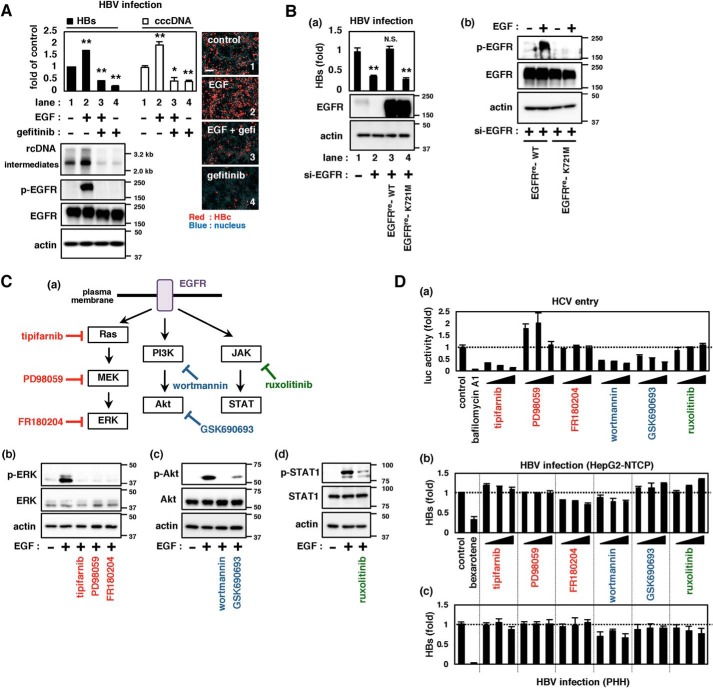
Figure 1.
Role of EGFR-downstream signaling in supporting HBV infection. A, HBV infection upon activation or inactivation of EGFR. EGF or gefitinib was applied during HBV inoculation of HepG2-NTCP cells for 16 h, and HBV infection was evaluated by detecting HBs in the culture supernatant (top left, black bars) as well as cccDNA (top left, white bars), HBV DNAs (bottom left, top panel, relaxed circular (rc) DNA and replicative intermediates (intermediates)), and HBc (right pictures, red) in the cells at 12 days post-inoculation. Total EGFR (EGFR) and EGFR phosphorylated at Tyr-1068 (p-EGFR) as well as actin as an internal control (actin) were detected by immunoblotting (bottom left). The numbers on the right of the panels indicate the size markers of DNA (kb, top panel) and protein (kDa, second, third, and bottom panels). Scale bars, 100 μm. B, ability of EGFR and its mutant to support HBV infection. HepG2-NTCP cells transfected with or without siRNA against EGFR (si-EGFR) and transduced with either EGFRre-WT or EGFRre-K721M by lentivirus vector were evaluated for HBV infection by detecting HBs in the culture supernatant (a, top graph). Protein expressions were also detected (a (middle and bottom panels) and b). C (a), schematic representations of the EGFR-downstream signaling (Ras-MAPK, PI3K-Akt, and JAK-STAT) and inhibitors for each cascade. b–d, effect of inhibitors on the target signaling. The indicated proteins were detected by immunoblot in HepG2-NTCP (b and c) or Huh-7 cells (d) treated with or without the indicated inhibitors. D, infection of HCV pseudoparticles to Huh-7 cells (a) and of HBV to HepG2-NTCP cells (b) and primary human hepatocytes (c) were evaluated upon treatment with or without the indicated inhibitors. Precise experimental procedures, including compound concentrations and treatment times, are shown under “Experimental procedures.” Error bars, S.D. Experiments were repeated three times (n = 3 independent experiments, each with triplicate samples). *, p < 0.05; **, p < 0.01; N.S., not significant.