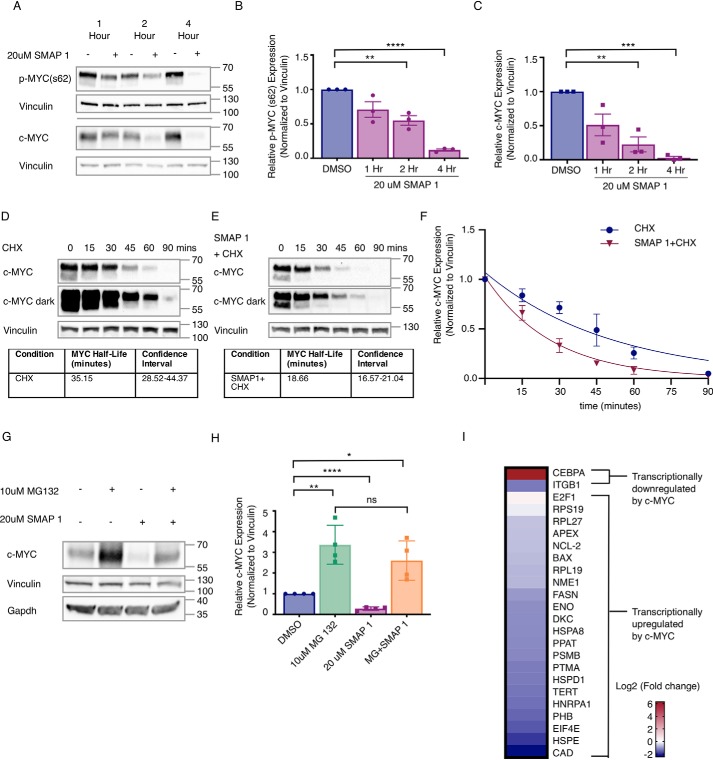
Figure 4.
SMAPs decrease c-MYC expression through a proteasome-mediated mechanism and induce changes to c-MYC target genes. A, Western blotting for Ser(P)62 and total c-MYC in Daudi cell line after exposure to 20 μm SMAP1 over 1, 2, or 4 h. B and C, quantification of phospho-MYC (B) and total c-MYC (C) protein in n = 3 experiments normalized to vinculin. D, Western blotting of MDA-MB-231 cells treated with cyclohexamide (CHX) over time and calculation of MYC t½. E, Western blotting for c-MYC expression in cells co-treated with SMAP1 and cyclohexamide over time and calculation of MYC t½. F, quantification of MYC in D and E over time used for calculation of c-MYC half-life in each condition and confidence interval. G, Western blotting for c-MYC expression upon treatment with MG132, SMAP1, or combination. H, quantification of G for n = 3. I, changes to mRNA expression of c-MYC target genes in Daudi cell line upon treatment with 20 μm SMAP1 for 6 h, representative of n = 4. c-MYC target genes are separated into two groups: up-regulated c-MYC target genes and down-regulated c-MYC target genes. Mean fold change, standard deviation, and p value are provided for each target in Table S2. Respective quantifications are represented as means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. ns, not significant.