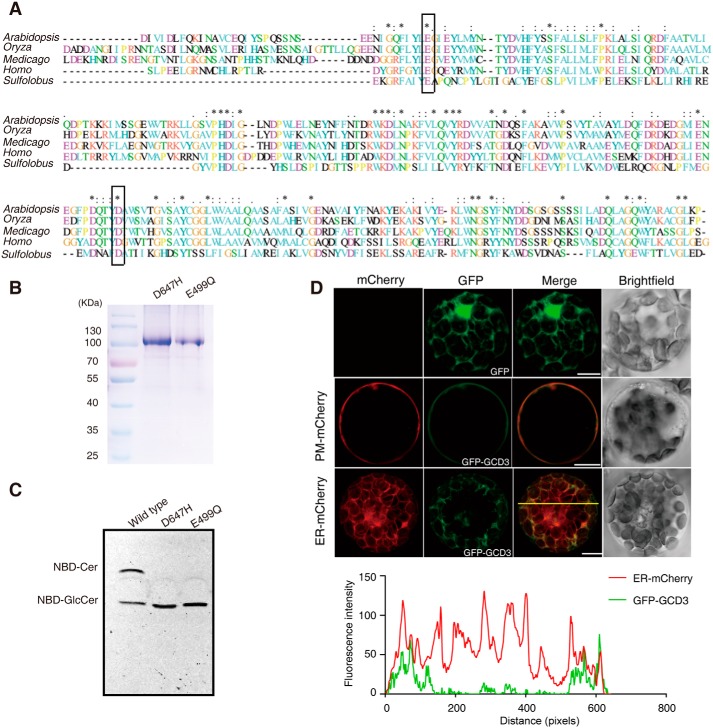
Figure 6.
Alignment of the AtGCD3 amino acid sequence with nonlysosomal glucosylceramidase and analysis of catalytic residues of AtGCD3. A, multiprotein alignment of the AtGCD3 amino acid sequence with other glucosylceramidases. Invariant residues are indicated with an asterisk; increased levels of conservation are indicated with a colon and a dot. The residues corresponding to the nucleophile Glu-499 and acid/base Asp-647 of AtGCD3 are boxed. Arabidopsis, gi|332657434 (AT4g10060) from A. thaliana; Oryza, gi|215704397 from Oryza sativa; Medicago, gi|355479745 from Medicago truncatula; Homo, NP_065995.1 from Homo sapiens; Sulfolobus, NC_002754.1 from S. solfataricus P2. B, SDS-PAGE of the purified E499Q and D647H mutant proteins. The two mutant enzymes were produced by site-directed mutagenesis, as shown in A (boxes), and were expressed and purified as described under “Experimental procedures.” C, TLC showing the hydrolysis of NBD-GlcCer by the WT and mutant AtGCD3. Note that AtGCD3 mutant proteins E499Q and D647H were completely inactive. D, subcellular localization of GFP-AtGCD3 fusion protein. GFP-AtGCD3 was co-transformed with organelle markers (mCherry) in WT Arabidopsis protoplasts. After incubation at 23 °C for 16 h under the light, fluorescence was observed by laser confocal microscopy. Row 3 panels are shown in three-dimensional maximum projection (eight sections at 0.84-μm step size), and others are single-plane. Free GFP was used as control (row 1 panels). The graph below shows the overlap of fluorescence intensity peaks along profiles as indicated in the merged micrograph. Bars, 10 μm.