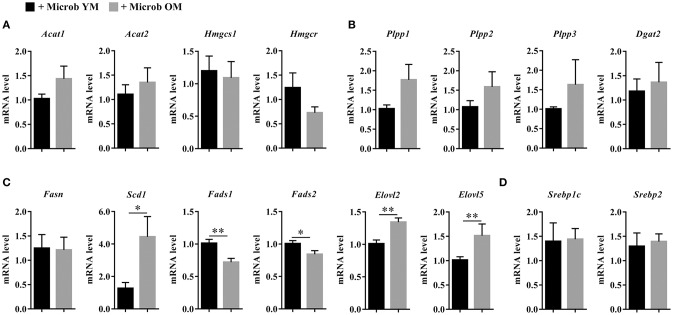
Figure 7.
Expression of hepatic genes involved in lipid biosynthesis. (A) Hepatic expression of genes encoding enzymes involved in the biosynthesis of cholesterol: acetyl-CoA acetyltransferase 1 and 2 (Acat1 and Acat2), hydroxymethylglutaryl-CoA synthase (Hmgcs1) and 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr). (B) Hepatic expression of genes encoding enzymes involved in the biosynthesis of di- and triglycerides: phospholipid phosphatases 1, 2, and 3 (Plpp1, Plpp2 and Plpp3) and diacylglycerol O-acyltransferase 2 (Dgat2). (C) Hepatic expression of genes encoding enzymes involved in the biosynthesis of fatty acids: fatty acid synthase (Fasn), acyl-CoA desaturase 1 (Scd1), acyl-CoA (8-3)-desaturase (Fads1), acyl-CoA 6-desaturase (Fads2), and elongation of very long chain fatty acids proteins 2 and 5 (Elovl2 and Elovl5). (D) Hepatic expression of genes encoding enzymes involved in the regulation of lipid synthesis: sterol regulatory element-binding proteins 1 (variant 3) and 2 (Srebp1c and Srebp2). The levels of mRNA were normalized to Hprt mRNA level for calculation of the relative levels of transcripts. mRNA levels are illustrated as fold change. All data are presented as mean ± SEM. n = 7 for the group of mice inoculated with the microbiota of young mice and n = 8 for the group of mice inoculated with the microbiota of old mice. Comparisons were done between the two groups of mice (+ Microb YM and + Microb OM). *p < 0.05 and **p < 0.01.