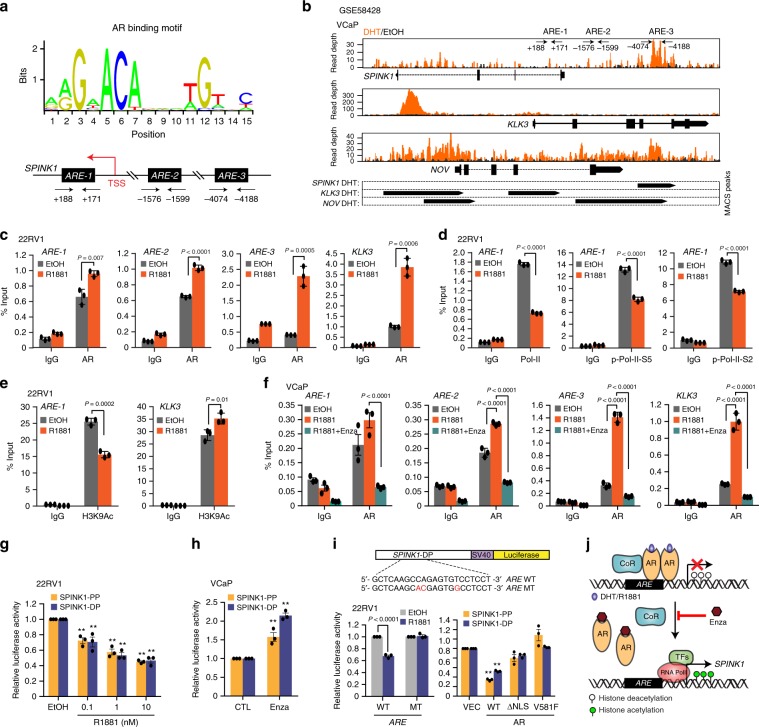
Fig. 3. AR directly binds to SPINK1 promoter region and modulates its expression.
a Schema showing AR binding motif obtained from JASPAR database (top). Bottom panel showing genomic location for the AREs on the SPINK1 promoter. b ChIP-Seq profiles indicating AR enrichment on the SPINK1, KLK3, and NOV gene loci in androgen stimulated VCaP cells (GSE58428). Bottom panel indicates the MACS identified peaks for AR binding on the promoters of SPINK1, KLK3, and NOV. c ChIP-qPCR data showing recruitment of AR on the SPINK1 promoter upon R1881 (10 nM) stimulation in 22RV1 cells. KLK3 promoter was used as a positive control for the androgen stimulation experiment. d Same as in c, except total RNA Pol-II, p-Pol-II-Ser5 and p-Pol-II-Ser2 on the SPINK1 promoter. e Same as in c, except H3K9Ac (H3 lysine 9 acetylation) marks on the SPINK1 and KLK3 promoters. f ChIP-qPCR data depicting enrichment of AR on the SPINK1 and KLK3 promoters in R1881 (10 nM) stimulated VCaP cells treated with or without enzalutamide (10 µM). g Luciferase reporter activity of the proximal (SPINK1-PP) and distal SPINK1 (SPINK1-DP) promoters in R1881(10 nM) stimulated 22RV1 cells. h Same as in g except enzalutamide (10 µM) treated VCaP cells were used. i Schematic showing luciferase reporter constructs with SPINK1-DP wild-type (WT) or mutated (MT) ARE sites (altered residues in red) (top). Bar plots showing luciferase reporter activity of SPINK1-DP WT or MT in R1881 stimulated (10 nM) 22RV1 cells (bottom, left) and 22RV1 cells co-transfected with SPINK1-PP or SPINK1-DP and control vector (VEC), AR wildtype (WT) or AR mutants (ΔNLS and V581F) constructs (bottom, right). j Illustration showing AR signaling mediated regulation of SPINK1 in prostate cancer, wherein CoR (corepressor), TFs (transcription factors) and Enza (enzalutamide) is shown. Experiments were performed with n = 3 biologically independent samples; data represents mean ± SEM. For panels c, d, e two-tailed unpaired Student’s t-test; g, i two-way ANOVA, Dunnett’s multiple-comparisons test; f, h two-way ANOVA, Sidak’s multiple-comparisons test was applied. ∗P ≤ 0.05 and ∗∗P ≤ 0.001.