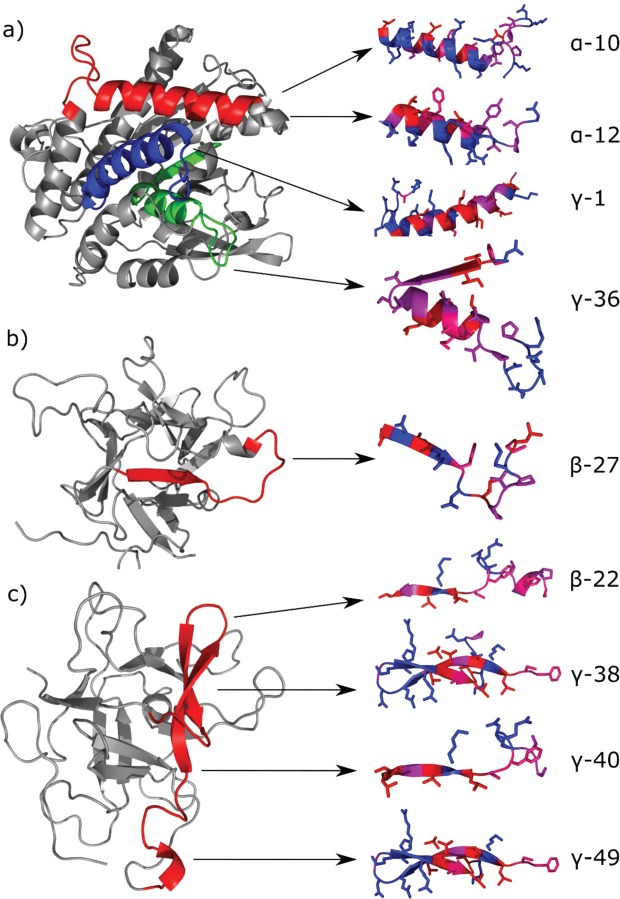
Figure 3.
Left: Localization of predicted emulsifier peptides based on template homology modelling using SWISS-MODEL Workspace39 (https://swissmodel.expasy.org/) and visualized in PyMOL version 1.5.0 (https://pymol.org/2/). Models are made using the protein of origin for the predicted peptides (Table 1). (a) Patatin-B2 (UniProt AC# P15477) was modelled using the X-ray structure of Patatin-17 (SMTL ID 4pka.1.A) as template, showing localization of the overlapping α-10 and α-12 (red), γ-1 (blue), and γ-36 (green). (b) KTI-A (UniProt AC# Q3S488) was modelled using the X-ray structure of Aspartic Protease inhibitor 11 (SMTL ID 5dzu.1.A) as template, showing localization of β-27 (red). (c) KTI-B (UniProt AC# Q3S474) was modelled using the X-ray structure of Kunitz-type protease inhibitor P1H5 (SMTL ID 3tc2.1.A) as template, showing localization of the overlapping β-22, γ-38, and γ-40 (red). The model for γ-49 (UniProt AC# Q3S477) using P1H5 (SMTL ID 3tc2.1.A) is superimposable with the model for the remaining KTI-B-derived peptides (i.e. γ-49 has the same structure) and is shown on the same model for simplicity. Right: Secondary structure of predicted peptides (as found within the models for their proteins of origin) with visible side chains and residue coloring according to the Swiss-Model hydrophobicity color scale (most hydrophobic residues in red and most hydrophilic residues in blue). Individually visualized proteins/peptides were assembled into the final figure using INKSCAPE version 0.92.3 (https://inkscape.org/).