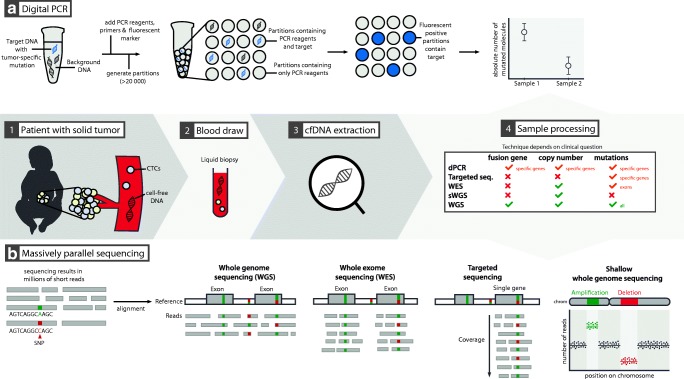
Fig. 1.
The optimal technique for cell-free DNA (cfDNA) analysis is chosen depending on the clinical question at hand. Commonly used techniques are digital PCR (dPCR) and massively parallel, or “next-generation,” sequencing. Digital PCR (panel a), with its unmatched sensitivity, is suited for monitoring known (hotspot) mutations, can be used to detect amplifications or losses of one or two pre-specified genomic regions and to detect pre-defined sites of genomic fusion. Massive parallel sequencing (panel b) is useful to detect all types of genomic alterations, depending on the sequencing strategy used. It can evaluate single nucleotide variants (SNVs), copy number aberrations (CNA), genomic fusions or a combination thereof. Whole-genome sequencing (WGS) results in uniform coverage across the entire genome. When performed at low coverage, the technique is termed shallow WGS, and is a cost-effective method to detect CNAs. Performed at higher coverage, the detailed analysis of mutations or translocations on a genome-wide scale is feasible. Whole-exome sequencing (WES) focuses the sequencing effort on the coding regions of the genome, but non-coding or structural variation is largely missed. Targeted sequencing will result in extremely high coverage over a small proportion of the genome, allowing the detection of variants in that specific region with high sensitivity. PCR, polymerase chain reaction. CTC, circulating tumor cells