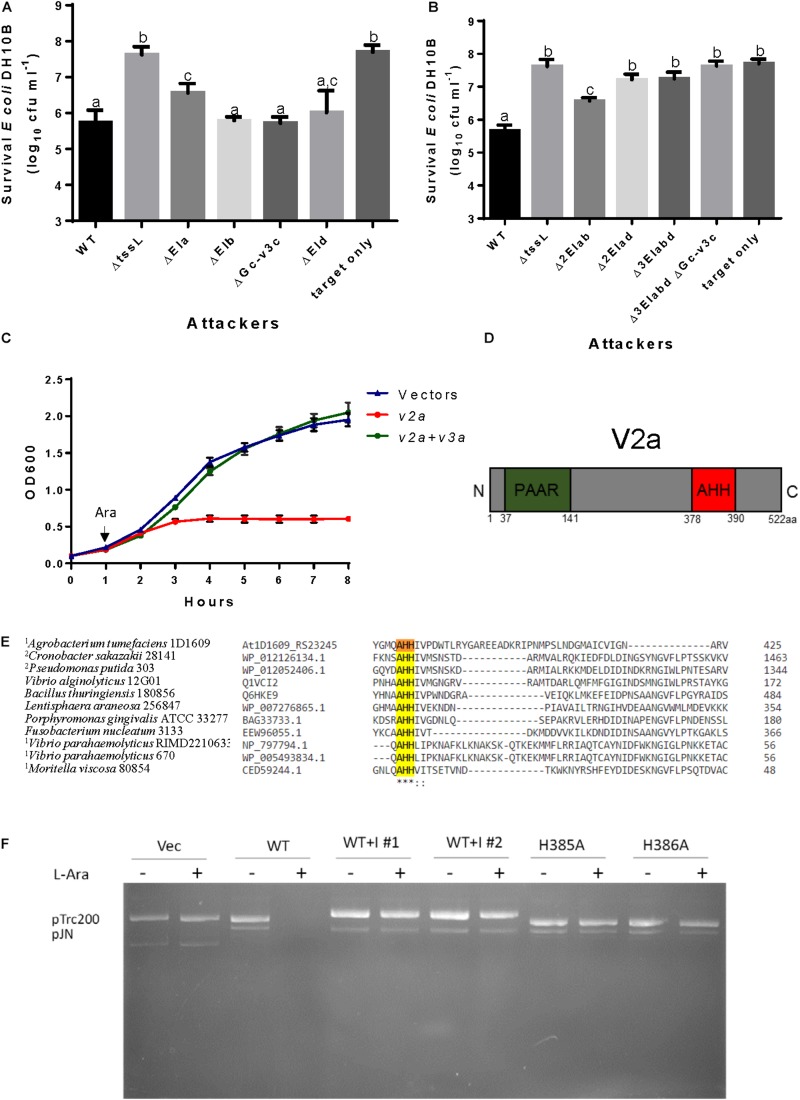
FIGURE 5.
vgrGa- and vgrGd-associated EI pairs are responsible for antibacterial activity and V2a is a nuclease. A. tumefaciens 1D1609 strains (indicated in the x axis) was mixed with E. coli DH10B cells expressing pRL662 to confer Gentamicin resistance at 30:1 ratio in LB. The survival of E. coli cells was quantified as cfu as shown on the y axis when co-cultured with single (A) and multiple (B) EI pair deletion mutant. Data are mean ± SEM of four biological repeats from two independent experiments. Different letters above the bar indicate statistically different groups of strains (P < 0.05) determined by Tukey’s HSD test, based on cfu of surviving E. coli cells. (C) E. coli growth inhibition analysis in vgrGa-associated EI pair. E. coli cultures were induced with 1 mM IPTG at 0 h for the expression of putative immunity protein expressed on pTrc200 plasmid followed by L-arabinose (Ara) induction at 1 hr to induce putative toxin gene from pJN105 plasmid. Cell growth was recorded every hour at OD600. Empty vectors were used as controls. Data are mean ± SEM of three independent experiments. (D) Schematic diagram showing N-terminal PAAR region and C-terminal AHH nuclease domain of vgrGa-associated effector V2a. (E) Partial sequence alignment of representative nuclease family of HNH/ENDO VII superfamily with conserved AHH motif, modified from NCBI CDD sequence alignment of nuclease family of the HNH/ENDO VII superfamily with conserved AHH (pfam14412). The strain name and locus tag/accession number are on the left, and the conserved amino acid residues are indicated as ∗ (identical) and: (similar) respectively. AHH motif is highlighted in yellow with the two His targeted for mutagenesis. AHH domain linked to DUF4150 domain1 or Rhs2 is indicted. (F) Nuclease activity assay. E. coli cells with pTrc200 and pJN105 plasmid (Vec) or derivatives expressing v2a (WT), WT with immunity protein (WT + I; #1 and #2 are two independent constructs) and catalytic site mutants (H385A, H386A) were induced with (+) or without (–) L-arabinose (L-Ara) for 2 h. Plasmid DNA was extracted and the degradation pattern was observed in agarose gel.