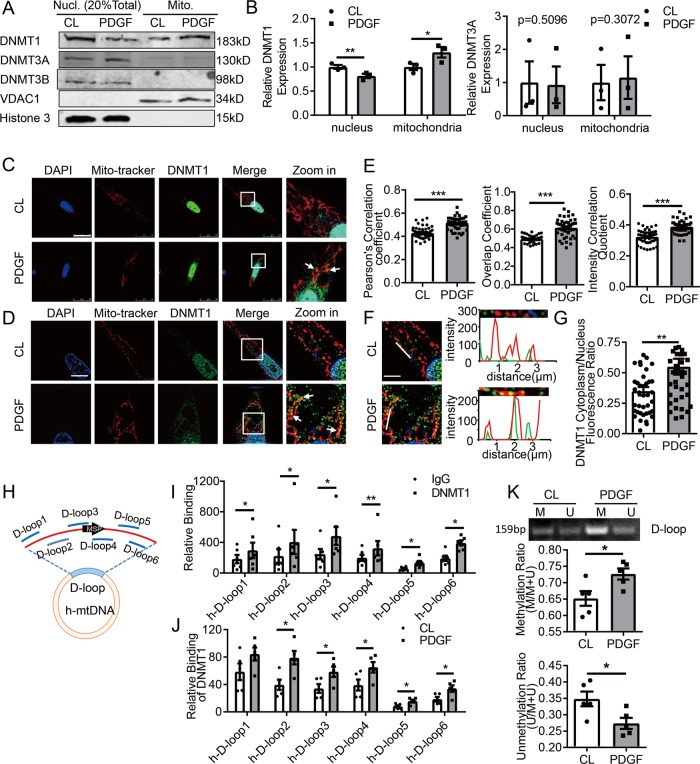
Fig. 1. DNA methyltransferase 1 (DNMT1) undergo a nucleus-to-mitochondrial translocation in PDGF-BB-stimulated vascular smooth muscle cells.
Vascular SMCs were subjected to vehicle (CL) or PDGF-BB (20 ng/mL) treatments for 24 h, and a accumulations of DNMT1, -3A, and -3B in nuclear (Nucl.) and mitochondrial (Mito.) fractions were analyzed (n = 3). Shown are representative blots. b Semi-quantification of DNMT1 and -3A in A. Expression was normalized to the respective internal reference proteins (VDAC1 or Histone 3) and expression in CL cells was set as 1. c, d Subcellular localization of DNMT1 was assessed by immunofluorescent staining followed by confocal microscopy (c) or structured illumination microscopy (d). Mito-tracker illustrates mitochondria. Arrowheads indicated co-localizations. Scale bar: 25 μm for c and 10 μm for d. e Co-localization of DNMT1 with mitochondria (mito-tracker) were quantified. Cells were randomly selected in microscopic fields from five biological replicates (n = 45). f Fluorescence intensity profiles indicate the degree of overlap between DNMT1 and mitochondria. Scale bar: 25 μm. g Ratio of fluorescence intensity of DNMT1 in cytoplasm over nucleus was measured (n = 40). h Schematic diagram of human mitochondrial DNA (h-mtDNA). Locations of primer sets for chromatin immunoprecipitation (ChIP) and methylation-specific PCR (MSP) were indicated. i DNMT1 bindings to D-loop regions in cells with CL treatment were analyzed by ChIP assay (n = 5). j DNMT1 bindings to D-loop regions in CL- and PDGF-BB-treated cells were analyzed (n = 5). k Methylation status of the D-loop region was measured by MSP (n = 5). M: methylated; U: unmethylated. *P < 0.05 **P < 0.01 ***P < 0.005 by student’s t-test (b, i, j, and k) and Mann–Whitney test (e, g), Error bars show ± SEM.