Figure 1.
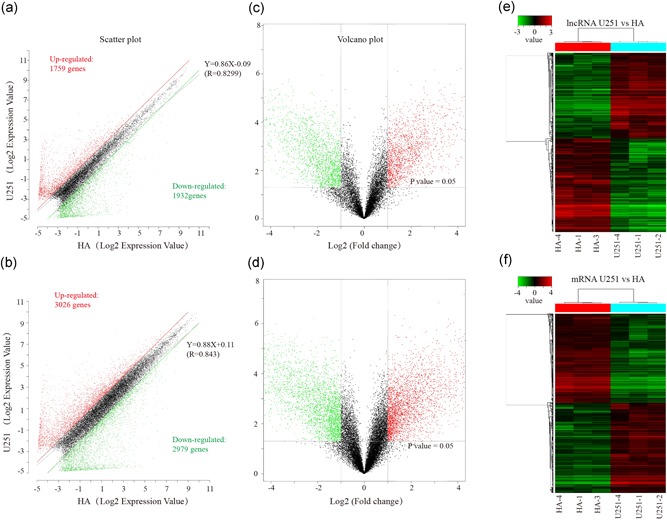
Expression profiles of lncRNAs and mRNAs in U251 glioma cells compared to HA samples. (a and b) Scatter‐plot of variation in expression of lncRNAs and mRNAs between U251 glioma cells and HA samples. X and Y axes are the mean normalized signal values (log2 scaled). The red and green lines are fold change lines (the default fold change value given is 2.0). (c and d) The Volcano plot of differentially expressed lncRNAs and mRNAs in U251 glioma cells relative to HA samples. The vertical lines represent 2.0‐fold changes up and down, and the horizontal line represents an FDR‐value of 0.05. The red and green points in the plot represent the differentially expressed lncRNAs with statistical significance. “Red” indicates high relative expression, and “green” indicates low relative expression. X‐axes are the fold change values (log2 scaled), Y axes are the FDR‐values (log2 scaled). (e and f) Hierarchical clustering analysis of lncRNAs and mRNAs. FDR, false discovery rate; HA, human astrocyte; lncRNA, long noncoding RNA; mRNA, messenger RNA
