Figure 1.
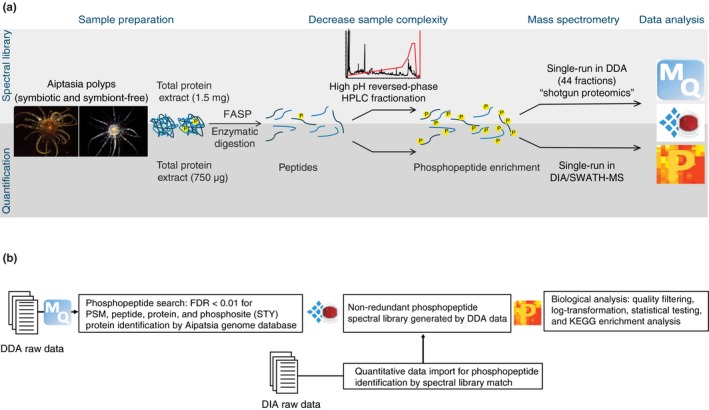
(a) Aiptasia sample preparation workflow for phosphoproteomics. Proteins were extracted from aposymbiotic and symbiotic Aiptasia polyps and digested in‐column. For spectral library generation, peptides were HPLC‐fractionated before phosphopeptide enrichment, and MS acquisition was performed in DDA mode. For phosphopeptide quantification no HPLC fractionation was performed and acquisition was performed in DIA/SWATH‐MS. (b) Data processing and analysis workflow. The DDA raw files were imported into MaxQuant for phosphopeptide search and used for spectral library generation in Spectronaut X. The quantitative DIA data were imported into Spectronaut X for retrieval of phosphopeptide identity by spectral library match. Biological and statistical data analysis was conducted in Perseus
