Figure 6.
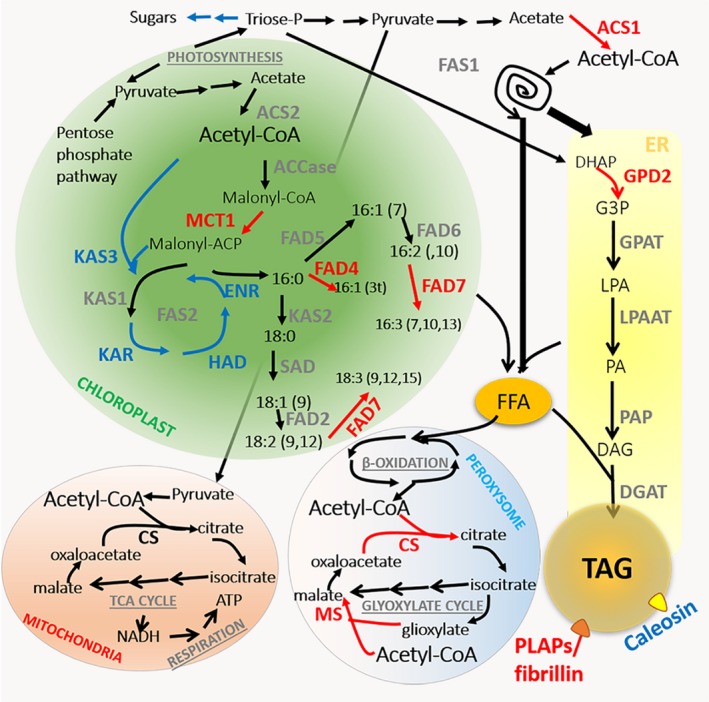
A schematic model of lipid biosynthesis in Chlorella vulgaris 211/11P. Proteins encoded by differently expressed genes are indicated in red (upregulated in HL) or in blue (downregulated in HL). Similar color code is used for chemical reactions catalyzed by differently expressed enzymes. ACCase, acetyl‐CoA carboxylase; MCT, malonyl‐CoA: ACP transacylase; KAS, 3‐ketoacyl‐ synthase; KAR, 3‐oxoacy‐ACP reductase; HAD, 3‐hydroxyacyl‐ACP dehydratase; ENR, enoyl‐ACP reductase; FAS1/2, fatty acid synthase type 1/2; SAD, stearate desaturase; FAD2, ω‐6 fatty acid desaturase, Δ12; FAD4, Δ3 palmitate desaturase; FAD5, palmitate Δ7 desaturase; FAD6, ω6 fatty acid desaturase; FAD7, chloroplast glycerolipid ω3 fatty acid desaturase; FFA, free fatty acids; CS, citrate synthase; MS, malate synthase ACS, acetyl‐CoA synthase; DHAP, dihydroxyacetone phosphate; GPD, glycerol‐3‐phosphate dehydrogenase; G3P, glycerol‐3‐phosphate; GPAT, Glycerol‐3‐phosphate O‐acyltransferase; LPA, lysophosphatidic acid; LPAAT, lysophosphatidic acid acyltransferase; PA, phosphatidic acid; PAP, phosphatidate phosphatase; DAG, diacylglycerol; DGAT, diacylglycerol acyltransferase; PLAPs/fibrillin, Plastid‐lipid‐associated protein PAP/fibrillin family protein; ER, endoplasmic reticulum; TAG, triacyclglycerols.
