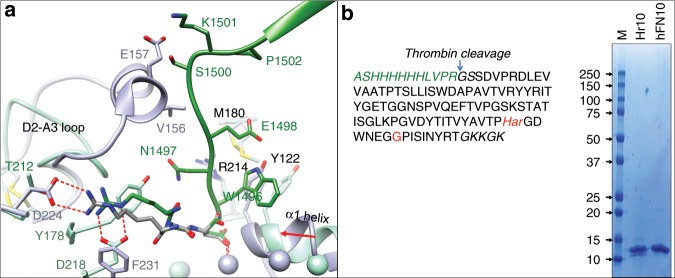
Fig. 1. Structure-guided design and binding properties of Hr10.
a Ribbon diagrams of the crystal structures of αVβ3/hFN10 (light green, 4mmz.pdb) and αIIbβ3/eptifibatide (light purple, 2vdn.pdb) superposed on the respective βA domains, with the metal ions at the ligand-associated metal-binding site (LIMBS), MIDAS, and adjacent to MIDAS (ADMIDAS) shown as spheres in the respective colors. Relevant segments of the propeller and βA domains and of hFN10 (dark green) and eptifibatide (dark gray) are shown. The MIDAS ion is ligated by the aspartate residue from each ligand. Residues (single letter code) specific to each structure are shown in the respective color, with residues or loops common in both shown in black. Oxygen, nitrogen, and sulfur atoms are in red, blue, and yellow, respectively. The inward movement (red arrow) of the α1 helix and ADMIDAS ion in αIIbβ3, driven by binding of the partial agonist eptifibatide, is absent in hFN10-bound αVβ3, the result of a π–π interaction between hFN10-W1496 and βA-Y122. βA-R214 and βA-M180 contribute to the stability of hFN10-W1496. Eptifibatide-Har2 forms a bidentate salt bridge with αIIb-D224, whereas hFN10-R1493 contacts αV-D218 (replaced by F231 in αIIb). b The translated sequence of His-tagged Hr10 lacking the N-terminal methionine. Har and glycine substitutions are indicated in red. Alien residues are in italics (green text). The isotopically averaged calculated molecular weight of Hr10 was 11,969.3 (protein calculator v3.4 http://protcalc.sourceforge.net/cgi-bin/protcalc). Inset, Coomassie stain of a whole 10–20% SDS–PAGE showing purified Hr10 and hFN10 (8 μg in each lane). MW markers (in kDa) are indicated.