Figure 1.
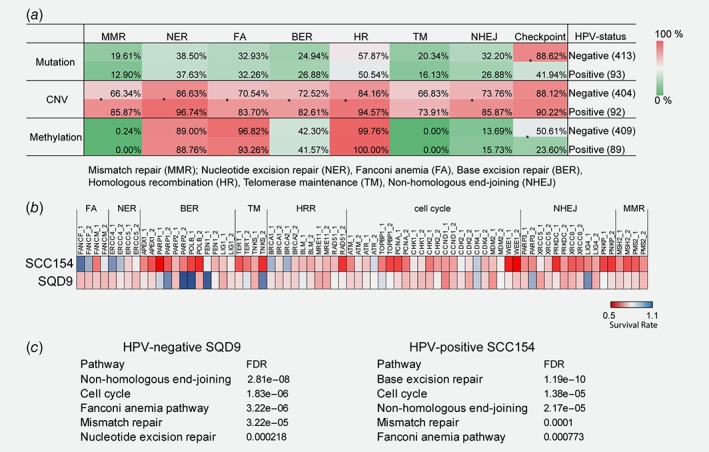
Genomic and functional differences in DDR of HPV‐positive and HPV‐negative HNSCCs. (a) Pathway enrichment analysis of the core DDR genes in the TCGA HNSCC samples. *p < 0.05 determined by Fisher's exact test. (b) CRISPR‐Cas9 screen using a library of 36 genes involved in DNA repair. A heatmap shows radiosensitization mediated by specific CRISPR‐Cas9 in SQD9 and SCC154 cells treated with 6 and 3 Gy, respectively. (c) KEGG pathway mapping of the screen hits analyzed by STRING. The order of enrichment is based on the false discovery rates (FDR).
