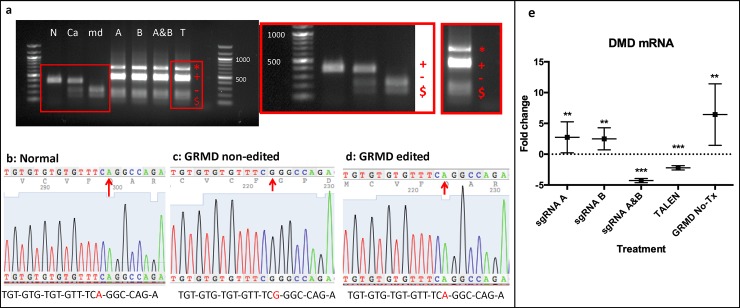
Fig 2. DNA and RNA analysis revealed HDR-mediated gene editing in GRMD-treated (Tx) myoblasts.
(a) Agarose gel after PCR and restriction digest of GRMD-Tx and non-Tx cells. From left to right: Normal (N), carrier (Ca), non-Tx GRMD (md), GRMD-sgRNA A-Tx (A), GRMD-sgRNA B-Tx (B), GRMD-sgRNA A&B combined Tx (A&B), GRMD-TALEN Tx (T), ladder (100bp). All bands were sequenced: top band of ~ 700bp (red asterisk) matched part of the sequence of the donor clone after being cut with Sau96I enzyme, second band of ~ 500bp (red cross) was the corrected DMD gene sequence in GRMD-HDR-Tx samples and normal dog cells. This band was not cut with Sau96I. This second band had a higher molecular weight in GRMD-Tx cells compared to normal due to additional genes (eGFP) present in the donor clone. Third and fourth bands ~ 200bp (red dash, red cash sign) correspond to fragments of the GRMD mutated dog genome that was cut with Sau96I. (b) Sanger sequencing from the cut band (red cross) in a normal dog. Red arrow denotes the correct bp (A) in the DMD gene. (c) Sanger sequencing from the cut band in GRMD-HDR-Tx myoblasts but not successfully edited. Red arrow points at mutated bp (G) in GRMD dogs (d) Sanger sequencing from cut band in successfully GRMD-HDR-Tx GRMD cells. Red arrow denotes successfully replaced bp (A). (e) Dystrophin mRNA expression (mean±SE) among HDR treatments and normal cells compared to normal cells expression. *** p ≤ 0.001, ** p ≤ 0.01 vs Normal. Samples were analyzed using a pair wise fixed reallocation randomization test, excluding outliers with a Grubb’s test. Vertical bars indicate standard error of the mean. Treated myoblasts were differentiated into myotubes for 18 to 21 days and RNA was extracted from 6 replicates; values were normalized to HPRT1 (house-keeping gene).