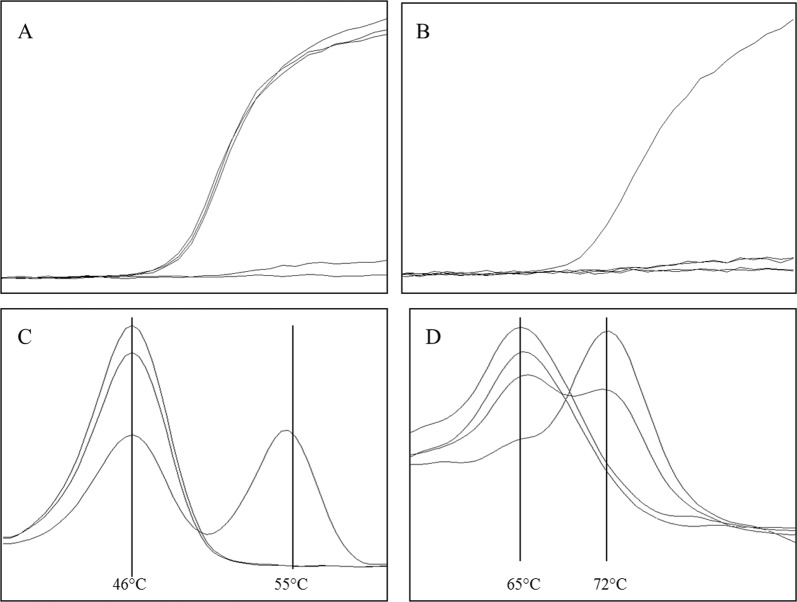
Fig. 1.
Multiplex PCR for simultaneously detection of CTNS variants. a Amplification signal for 57 kb deletion, detection filter 533-610, three samples show an exponential amplification signal, demonstrating the presence of the deletion. One sample was negative in this assay and the last one represents the negative control, both without amplification signal. No differentiation between homo- or heterozygous state is possible. b Amplification of parts of exon 3 used for differentiation between a homo- or heterozygous 57 kb deletion as well as amplification control (Channel 533–610). One sample gives an exponential amplification signal, indicating the presence of the wildtype region used for differentiation, two samples were negative and one is the negative control. c Detection of rs786204501 (c.18_21del GACT) via melting analysis (Channel 498–640). The mutated allele gives a melting peak of app. 55 °C, whereas the wildtype allele shows a peak of app. 46 °C. One sample is heterozygous for this variant, and the two other samples show the wildtype allele. d Detection of rs786204420 (c.926dupG) via melting analysis (Channel 465–510). Wildtype allele gives a melting peak at app. 65 °C and the insertion at app. 72 °C. Two samples are wildtype for the tested variant, with a melting point at 65 °C, one sample is homozygous for the c.926dupG giving a melting peak at 72 °C and one sample is heterozygous for this variant